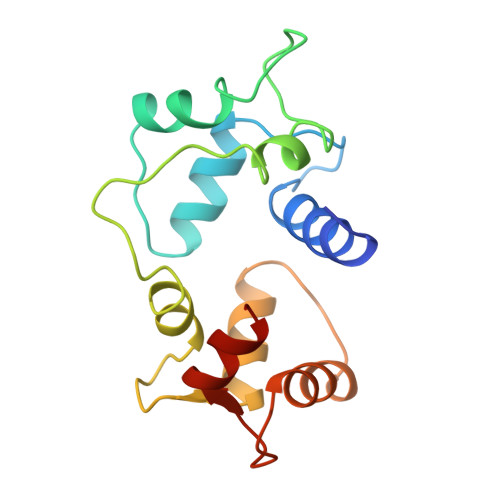
Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Wall, M.E., Clarage, J.B., Phillips Jr., G.N.(1997) Structure 5: 1599-1612
- PubMed: 9438860
- DOI: https://doi.org/10.1016/s0969-2126(97)00308-0
- Primary Citation of Related Structures:
1CM1, 1CM4 - PubMed Abstract:
Calmodulin is a calcium-activated regulatory protein which can bind to many different targets. The protein resembles a highly flexible dumbbell, and bends in the middle as it binds. This and other motions must be understood to formulate a realistic model of calmodulin function.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, The WM Keck Center for Computational Biology, Rice University Houston, TX 77005-1892, USA, mewall@bioc.rice.edu