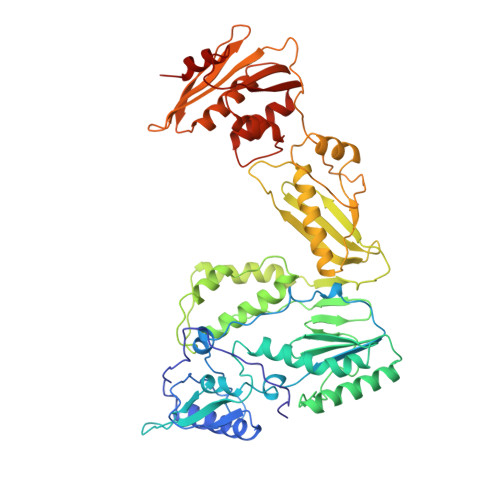
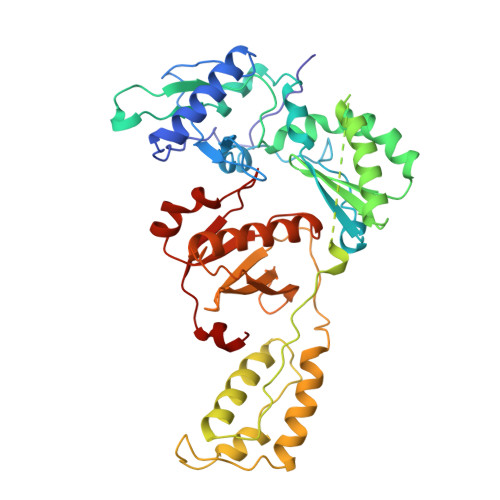
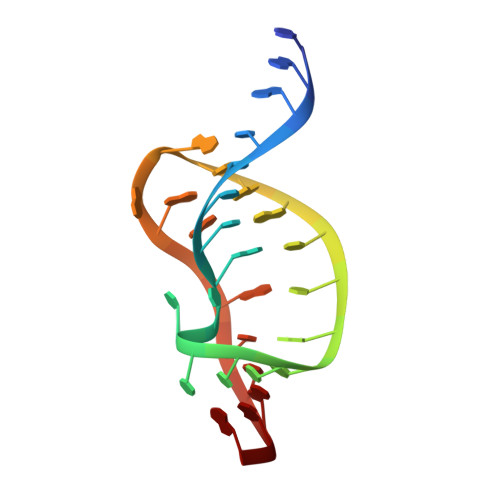
The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor.
Jaeger, J., Restle, T., Steitz, T.A.(1998) EMBO J 17: 4535-4542
- PubMed: 9687519
- DOI: https://doi.org/10.1093/emboj/17.15.4535
- Primary Citation of Related Structures:
1HVU - PubMed Abstract:
Small RNA pseudoknots, selected to bind human immunodeficiency virus type 1 (HIV-1) reverse transcriptase tightly, are potent inhibitors of reverse transcriptase. The co-crystal structure of reverse transcriptase complexed with a 33 nucleotide RNA pseudoknot has been determined by fitting the ligand into a high quality, 4-fold averaged 4.8 A resolution electron density map. The RNA is kinked between stems S1 and S2, thereby optimizing its contacts with subunits of the heterodimer. Its binding site extends along the cleft that lies between the polymerase and RNase H active sites, partially overlaps with that observed for duplex DNA and presumably overlaps some portion of the tRNA site. Stem S2 and loop L1 stabilize the 'closed' conformation of the polymerase through extensive electrostatic interactions with several basic residues in helix I of the p66 thumb and in the p66 fingers domain. Presumably, this RNA ligand inhibits reverse transcriptase by binding to a site that partly overlaps the primer-template binding site.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114, USA.