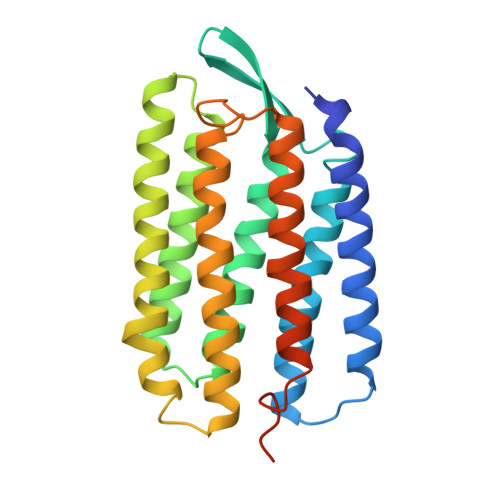
Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Belrhali, H., Nollert, P., Royant, A., Menzel, C., Rosenbusch, J.P., Landau, E.M., Pebay-Peyroula, E.(1999) Structure 7: 909-917
- PubMed: 10467143
- DOI: https://doi.org/10.1016/s0969-2126(99)80118-x
- Primary Citation of Related Structures:
1QHJ - PubMed Abstract:
Bacteriorhodopsin (bR) from Halobacterium salinarum is a proton pump that converts the energy of light into a proton gradient that drives ATP synthesis. The protein comprises seven transmembrane helices and in vivo is organized into purple patches, in which bR and lipids form a crystalline two-dimensional array. Upon absorption of a photon, retinal, which is covalently bound to Lys216 via a Schiff base, is isomerized to a 13-cis,15-anti configuration. This initiates a sequence of events - the photocycle - during which a proton is transferred from the Schiff base to Asp85, followed by proton release into the extracellular medium and reprotonation from the cytoplasmic side.
Organizational Affiliation:
ESRF, Grenoble, France.