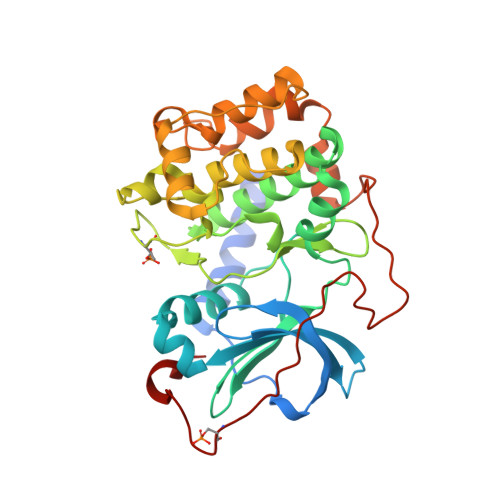
The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
Jacobs, M., Hayakawa, K., Swenson, L., Bellon, S., Fleming, M., Taslimi, P., Doran, J.(2006) J Biol Chem 281: 260-268
- PubMed: 16249185
- DOI: https://doi.org/10.1074/jbc.M508847200
- Primary Citation of Related Structures:
2ERZ, 2ESM, 2ETK, 2ETR, 3D9V - PubMed Abstract:
ROCK or Rho-associated kinase, a serine/threonine kinase, is an effector of Rho-dependent signaling and is involved in actin-cytoskeleton assembly and cell motility and contraction. The ROCK protein consists of several domains: an N-terminal region, a kinase catalytic domain, a coiled-coil domain containing a RhoA binding site, and a pleckstrin homology domain. The C-terminal region of ROCK binds to and inhibits the kinase catalytic domains, and this inhibition is reversed by binding RhoA, a small GTPase. Here we present the structure of the N-terminal region and the kinase domain. In our structure, two N-terminal regions interact to form a dimerization domain linking two kinase domains together. This spatial arrangement presents the kinase active sites and regulatory sequences on a common face affording the possibility of both kinases simultaneously interacting with a dimeric inhibitory domain or with a dimeric substrate. The kinase domain adopts a catalytically competent conformation; however, no phosphorylation of active site residues is observed in the structure. We also determined the structures of ROCK bound to four different ATP-competitive small molecule inhibitors (Y-27632, fasudil, hydroxyfasudil, and H-1152P). Each of these compounds binds with reduced affinity to cAMP-dependent kinase (PKA), a highly homologous kinase. Subtle differences exist between the ROCK- and PKA-bound conformations of the inhibitors that suggest that interactions with a single amino acid of the active site (Ala215 in ROCK and Thr183 in PKA) determine the relative selectivity of these compounds. Hydroxyfasudil, a metabolite of fasudil, may be selective for ROCK over PKA through a reversed binding orientation.
Organizational Affiliation:
Vertex Pharmaceuticals Incorporated, Cambridge, Massachusetts 02139, USA. marc_jacobs@vrtx.com