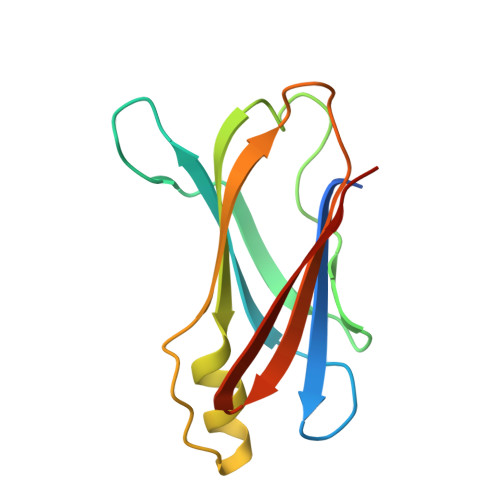
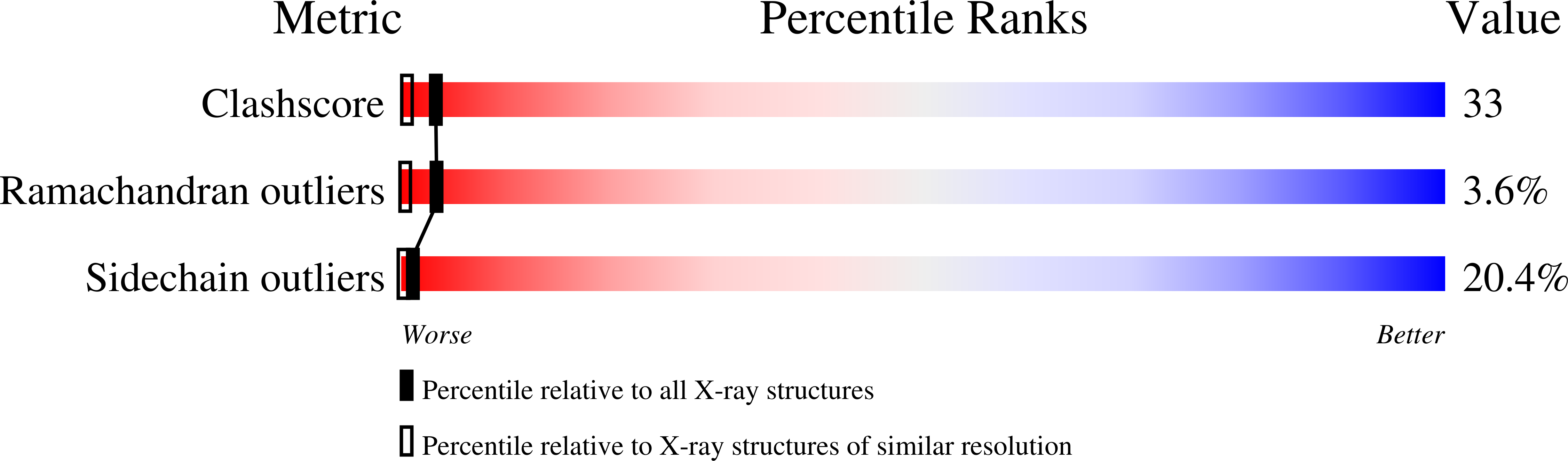Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A.
Blake, C.C., Geisow, M.J., Oatley, S.J., Rerat, B., Rerat, C.(1978) J Mol Biol 121: 339-356
- PubMed: 671542
- DOI: https://doi.org/10.1016/0022-2836(78)90368-6
- Primary Citation of Related Structures:
2PAB