Triazole derivatives as non-nucleoside inhibitors of HIV-1 reverse transcriptase-structure-activity relationships and crystallographic analysis.
Kirschberg, T.A., Balakrishnan, M., Huang, W., Hluhanich, R., Kutty, N., Liclican, A.C., McColl, D.J., Squires, N.H., Lansdon, E.B.(2008) Bioorg Med Chem Lett 18: 1131-1134
- PubMed: 18083512
- DOI: https://doi.org/10.1016/j.bmcl.2007.11.127
- Primary Citation of Related Structures:
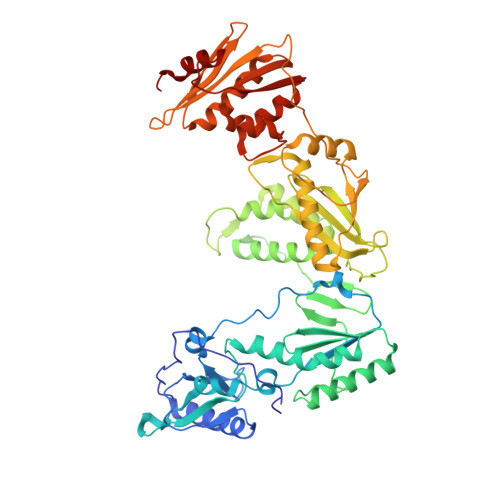
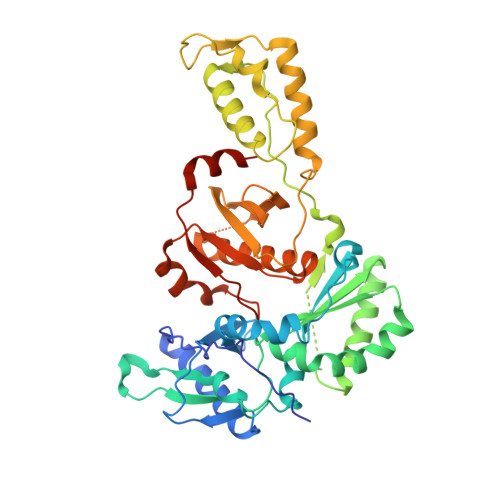
2RKI - PubMed Abstract:
A series of 3,4,5-trisubstituted 1,2,4-4H triazole derivatives was synthesized and investigated for HIV-1 reverse transcriptase inhibition. An X-ray structure with HIV-1 RT secured the binding mode and allowed the key interactions with the enzyme to be identified.
Organizational Affiliation:
Gilead Sciences, Department of Medicinal Chemistry, 333 Lakeside Drive, Foster City, CA 94404, USA. tkirschberg@gilead.com