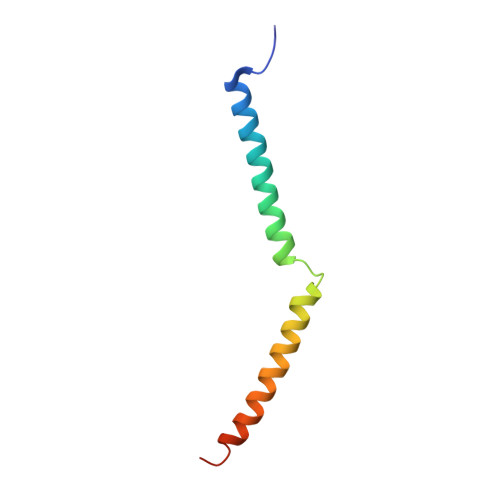
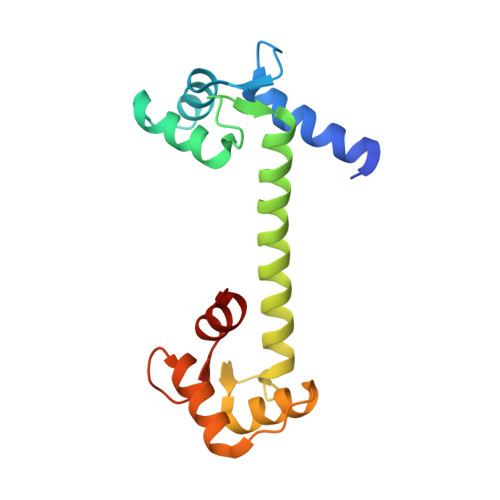
Structure of Human Na+/H+ Exchanger Nhe1 Regulatory Region in Complex with Cam and Ca2+
Koester, S., Pavkov-Keller, T., Kuehlbrandt, W., Yildiz, O.(2011) J Biol Chem 286: 40954
- PubMed: 21931166
- DOI: https://doi.org/10.1074/jbc.M111.286906
- Primary Citation of Related Structures:
2YGG - PubMed Abstract:
The ubiquitous mammalian Na(+)/H(+) exchanger NHE1 has critical functions in regulating intracellular pH, salt concentration, and cellular volume. The regulatory C-terminal domain of NHE1 is linked to the ion-translocating N-terminal membrane domain and acts as a scaffold for signaling complexes. A major interaction partner is calmodulin (CaM), which binds to two neighboring regions of NHE1 in a strongly Ca(2+)-dependent manner. Upon CaM binding, NHE1 is activated by a shift in sensitivity toward alkaline intracellular pH. Here we report the 2.23 Å crystal structure of the NHE1 CaM binding region (NHE1(CaMBR)) in complex with CaM and Ca(2+). The C- and N-lobes of CaM bind the first and second helix of NHE1(CaMBR), respectively. Both the NHE1 helices and the Ca(2+)-bound CaM are elongated, as confirmed by small angle x-ray scattering analysis. Our x-ray structure sheds new light on the molecular mechanisms of the phosphorylation-dependent regulation of NHE1 and enables us to propose a model of how Ca(2+) regulates NHE1 activity.
Organizational Affiliation:
Department of Structural Biology, Max Planck Institute of Biophysics, Max-von-Laue Str. 3, 60438 Frankfurt am Main, Germany.