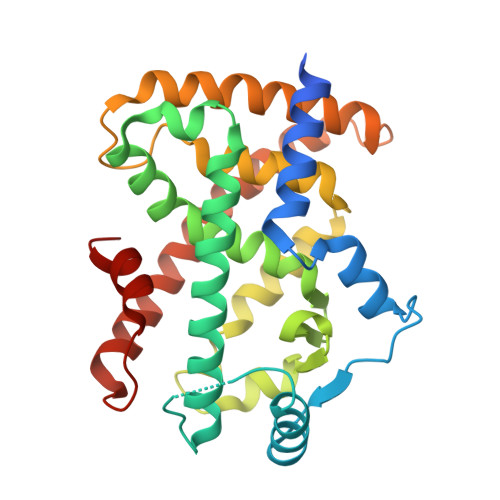
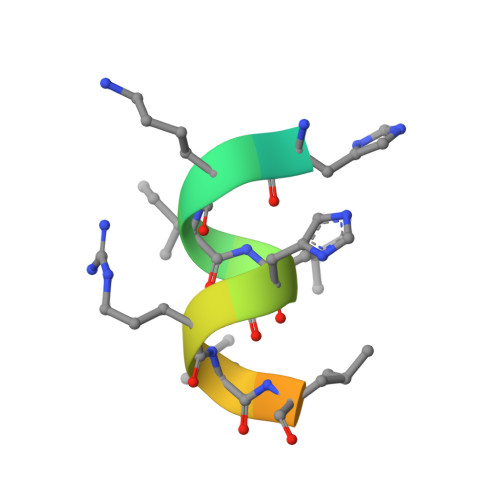
GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
Amato, A.A., Rajagopalan, S., Lin, J.Z., Carvalho, B.M., Figueira, A.C., Lu, J., Ayers, S.D., Mottin, M., Silveira, R.L., Souza, P.C., Mourao, R.H., Saad, M.J., Togashi, M., Simeoni, L.A., Abdalla, D.S., Skaf, M.S., Polikparpov, I., Lima, M.C., Galdino, S.L., Brennan, R.G., Baxter, J.D., Pitta, I.R., Webb, P., Phillips, K.J., Neves, F.A.(2012) J Biol Chem 287: 28169-28179
- PubMed: 22584573
- DOI: https://doi.org/10.1074/jbc.M111.332106
- Primary Citation of Related Structures:
3T03 - PubMed Abstract:
The recent discovery that peroxisome proliferator-activated receptor γ (PPARγ) targeted anti-diabetic drugs function by inhibiting Cdk5-mediated phosphorylation of the receptor has provided a new viewpoint to evaluate and perhaps develop improved insulin-sensitizing agents. Herein we report the development of a novel thiazolidinedione that retains similar anti-diabetic efficacy as rosiglitazone in mice yet does not elicit weight gain or edema, common side effects associated with full PPARγ activation. Further characterization of this compound shows GQ-16 to be an effective inhibitor of Cdk5-mediated phosphorylation of PPARγ. The structure of GQ-16 bound to PPARγ demonstrates that the compound utilizes a binding mode distinct from other reported PPARγ ligands, although it does share some structural features with other partial agonists, such as MRL-24 and PA-082, that have similarly been reported to dissociate insulin sensitization from weight gain. Hydrogen/deuterium exchange studies reveal that GQ-16 strongly stabilizes the β-sheet region of the receptor, presumably explaining the compound's efficacy in inhibiting Cdk5-mediated phosphorylation of Ser-273. Molecular dynamics simulations suggest that the partial agonist activity of GQ-16 results from the compound's weak ability to stabilize helix 12 in its active conformation. Our results suggest that the emerging model, whereby "ideal" PPARγ-based therapeutics stabilize the β-sheet/Ser-273 region and inhibit Cdk5-mediated phosphorylation while minimally invoking adipogenesis and classical agonism, is indeed a valid framework to develop improved PPARγ modulators that retain antidiabetic actions while minimizing untoward effects.
Organizational Affiliation:
Laboratório de Farmacologia Molecular, Departamento de Ciências Farmacêuticas, Faculdade de Ciências da Saúde, Universidade de Brasília, 70919-970 Brazil.