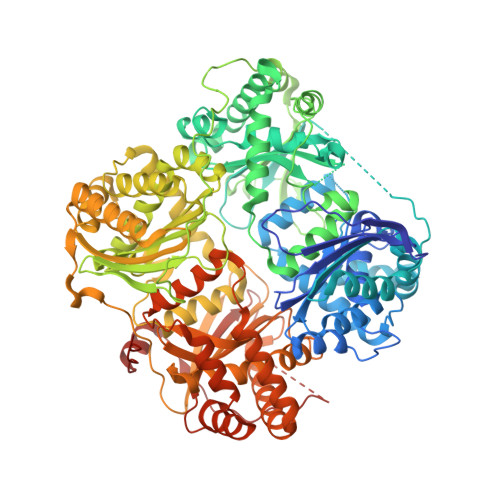
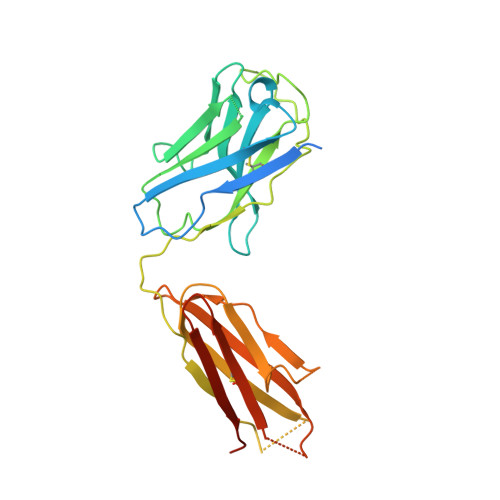
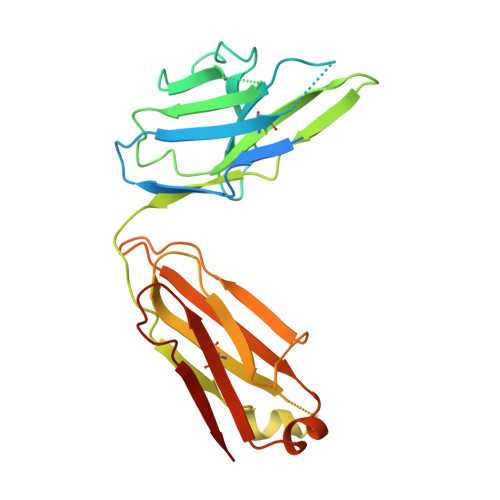
Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme.
McCord, L.A., Liang, W.G., Dowdell, E., Kalas, V., Hoey, R.J., Koide, A., Koide, S., Tang, W.J.(2013) Proc Natl Acad Sci U S A 110: 13827-13832
- PubMed: 23922390
- DOI: https://doi.org/10.1073/pnas.1304575110
- Primary Citation of Related Structures:
4IOF - PubMed Abstract:
Insulin-degrading enzyme (IDE) selectively degrades the monomer of amyloidogenic peptides and contributes to clearance of amyloid β (Aβ). Thus, IDE retards the progression of Alzheimer's disease. IDE possesses an enclosed catalytic chamber that engulfs and degrades its peptide substrates; however, the molecular mechanism of IDE function, including substrate access to the chamber and recognition, remains elusive. Here, we captured a unique IDE conformation by using a synthetic antibody fragment as a crystallization chaperone. An unexpected displacement of a door subdomain creates an ~18-Å opening to the chamber. This swinging-door mechanism permits the entry of short peptides into the catalytic chamber and disrupts the catalytic site within IDE door subdomain. Given the propensity of amyloidogenic peptides to convert into β-strands for their polymerization into amyloid fibrils, they also use such β-strands to stabilize the disrupted catalytic site resided at IDE door subdomain for their degradation by IDE. Thus, action of the swinging door allows IDE to recognize amyloidogenicity by substrate-induced stabilization of the IDE catalytic cleft. Small angle X-ray scattering (SAXS) analysis revealed that IDE exists as a mixture of closed and open states. These open states, which are distinct from the swinging door state, permit entry of larger substrates (e.g., Aβ, insulin) to the chamber and are preferred in solution. Mutational studies confirmed the critical roles of the door subdomain and hinge loop joining the N- and C-terminal halves of IDE for catalysis. Together, our data provide insights into the conformational changes of IDE that govern the selective destruction of amyloidogenic peptides.
Organizational Affiliation:
Ben-May Department for Cancer Research, University of Chicago, Chicago, IL 60637, USA.