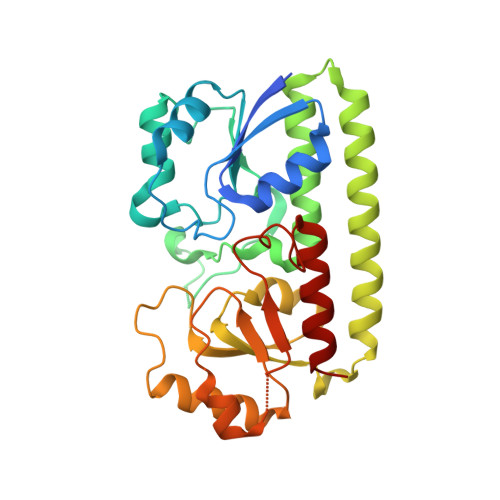
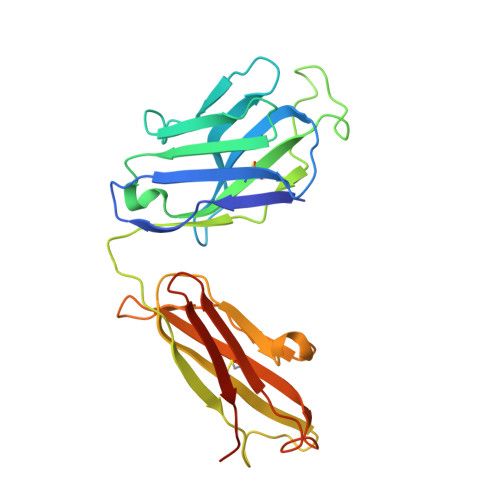
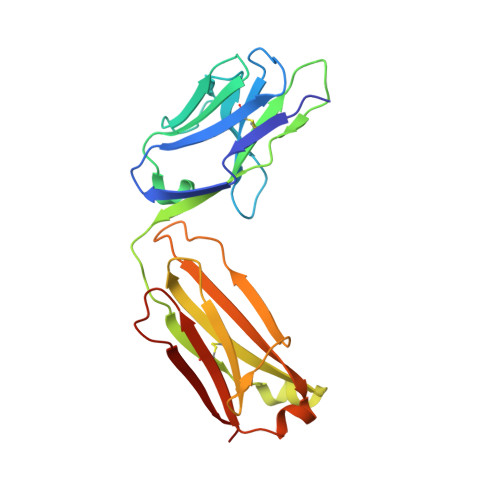
Structural analysis of bacterial ABC transporter inhibition by an antibody fragment.
Ahuja, S., Rouge, L., Swem, D.L., Sudhamsu, J., Wu, P., Russell, S.J., Alexander, M.K., Tam, C., Nishiyama, M., Starovasnik, M.A., Koth, C.M.(2015) Structure 23: 713-723
- PubMed: 25752540
- DOI: https://doi.org/10.1016/j.str.2015.01.020
- Primary Citation of Related Structures:
4NNO, 4NNP - PubMed Abstract:
Bacterial ATP-binding cassette (ABC) importers play critical roles in nutrient acquisition and are potential antibacterial targets. However, structural bases for their inhibition are poorly defined. These pathways typically rely on substrate binding proteins (SBPs), which are essential for substrate recognition, delivery, and transporter function. We report the crystal structure of a Staphylococcus aureus SBP for Mn(II), termed MntC, in complex with FabC1, a potent antibody inhibitor of the MntABC pathway. This pathway is essential and highly expressed during S. aureus infection and facilitates the import of Mn(II), a critical cofactor for enzymes that detoxify reactive oxygen species (ROS). Structure-based functional studies indicate that FabC1 sterically blocks a structurally conserved surface of MntC, preventing its interaction with the MntB membrane importer and increasing wild-type S. aureus sensitivity to oxidative stress by more than 10-fold. The results define an SBP blocking mechanism as the basis for ABC importer inhibition by an engineered antibody fragment.
Organizational Affiliation:
Department of Structural Biology, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080, USA.