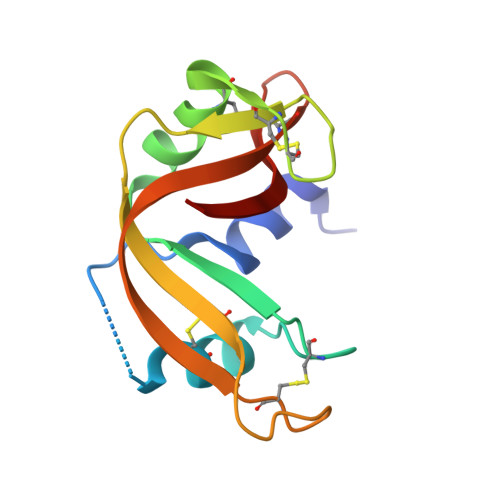
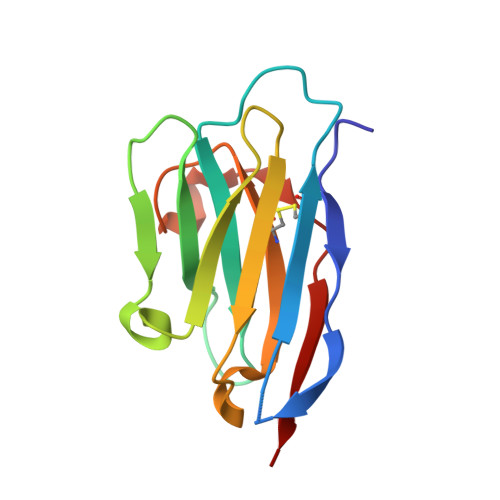
Structural basis of an engineered dual-specific antibody: conformational diversity leads to a hypervariable loop metal-binding site.
Fanning, S.W., Walter, R., Horn, J.R.(2014) Protein Eng Des Sel 27: 391-397
- PubMed: 25143596
- DOI: https://doi.org/10.1093/protein/gzu033
- Primary Citation of Related Structures:
4POU, 4POY, 4PPT - PubMed Abstract:
To explore dual-specificity in a small protein interface, we previously generated a 'metal switch' anti-RNase A VHH antibody using a combinatorial histidine library approach. While most metal-binding sites in proteins are found within rigid secondary structure, the engineered VHH antibody (VHH(metal)), which contained three new histidine residues, possessed metal-binding residues within the flexible hypervariable loops. Here, crystal structure analysis of the free and bound states of VHH(metal) reveals the structural determinants leading to dual-function. Most notably, CDR1 is observed in two distinct conformations when adopting the metal or RNase A bound states. Furthermore, mutagenesis studies revealed that one of the engineered residues, not located in the metal-binding pocket, contributed indirectly to metal recognition, likely through influencing CDR1 conformation. Despite these changes, VHH(metal) possesses a relatively minor energetic penalty toward binding the original antigen, RNase A (~1 kcal/mol), where the engineered gain-of-function metal-binding residues are observed to possess a mix of favorable and unfavorable contributions towards RNase A recognition. Ultimately, the conformationally distinct metal-switch interface architecture reflects the robust, library-based strategy used to produce VHH(metal). These results also suggest that even small protein interfaces, such as VHH, may be structurally and energetically forgiving in adopting novel function, while maintaining original function.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Northern Illinois University, DeKalb, IL 60115, USA Current address: Ben May Department for Cancer Research, University of Chicago, 929 E. 57th St., Chicago, IL 60637, USA.