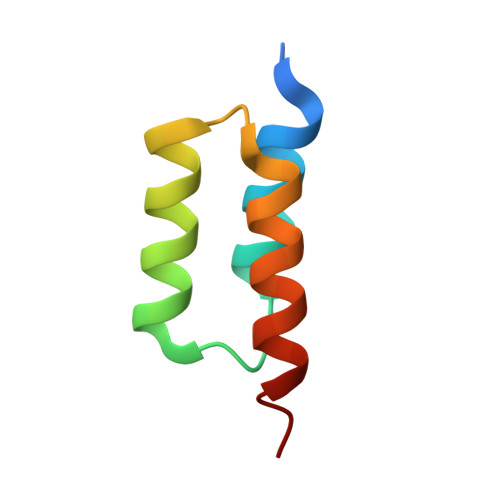
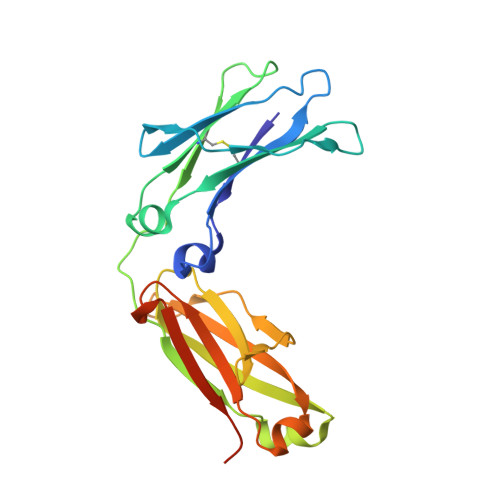
Suppression of conformational heterogeneity at a protein-protein interface.
Deis, L.N., Wu, Q., Wang, Y., Qi, Y., Daniels, K.G., Zhou, P., Oas, T.G.(2015) Proc Natl Acad Sci U S A 112: 9028-9033
- PubMed: 26157136
- DOI: https://doi.org/10.1073/pnas.1424724112
- Primary Citation of Related Structures:
4WWI, 4ZMD, 4ZNC - PubMed Abstract:
Staphylococcal protein A (SpA) is an important virulence factor from Staphylococcus aureus responsible for the bacterium's evasion of the host immune system. SpA includes five small three-helix-bundle domains that can each bind with high affinity to many host proteins such as antibodies. The interaction between a SpA domain and the Fc fragment of IgG was partially elucidated previously in the crystal structure 1FC2. Although informative, the previous structure was not properly folded and left many substantial questions unanswered, such as a detailed description of the tertiary structure of SpA domains in complex with Fc and the structural changes that take place upon binding. Here we report the 2.3-Å structure of a fully folded SpA domain in complex with Fc. Our structure indicates that there are extensive structural rearrangements necessary for binding Fc, including a general reduction in SpA conformational heterogeneity, freezing out of polyrotameric interfacial residues, and displacement of a SpA side chain by an Fc side chain in a molecular-recognition pocket. Such a loss of conformational heterogeneity upon formation of the protein-protein interface may occur when SpA binds its multiple binding partners. Suppression of conformational heterogeneity may be an important structural paradigm in functionally plastic proteins.
Organizational Affiliation:
Department of Biochemistry, Duke University, Durham, NC 27710.