Macyranones: Structure, Biosynthesis, and Binding Mode of an Unprecedented Epoxyketone that Targets the 20S Proteasome.
Keller, L., Plaza, A., Dubiella, C., Groll, M., Kaiser, M., Muller, R.(2015) J Am Chem Soc 137: 8121
- PubMed: 26050527
- DOI: https://doi.org/10.1021/jacs.5b03833
- Primary Citation of Related Structures:
5AHJ - PubMed Abstract:
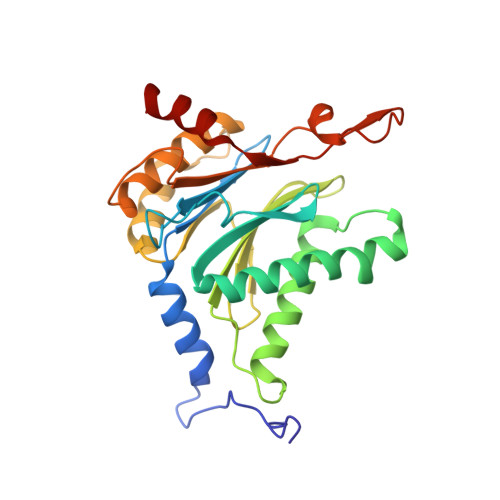
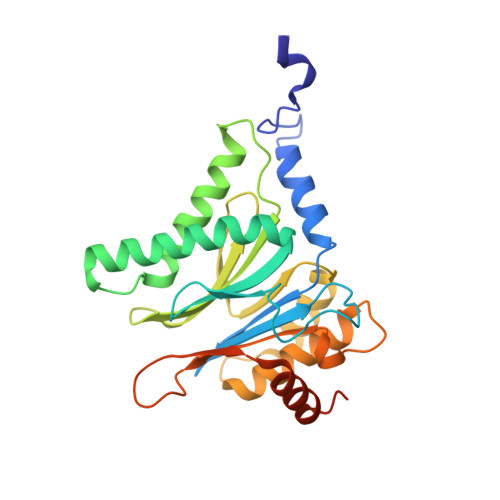
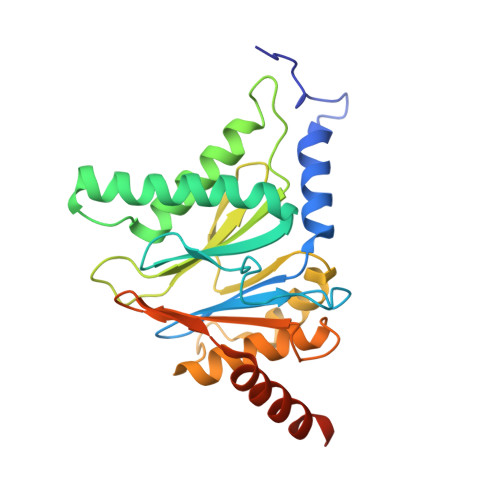
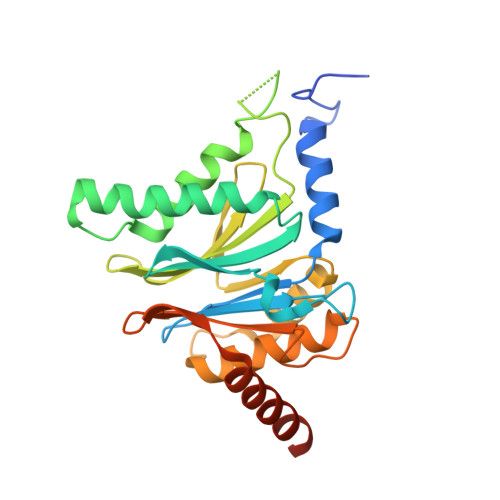
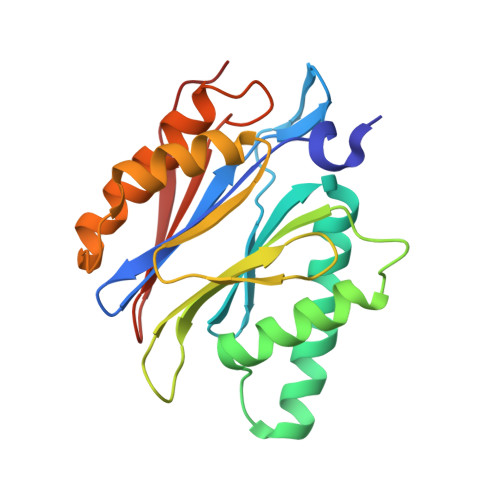
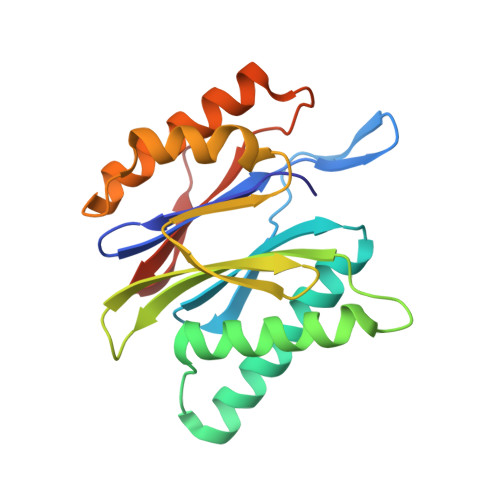
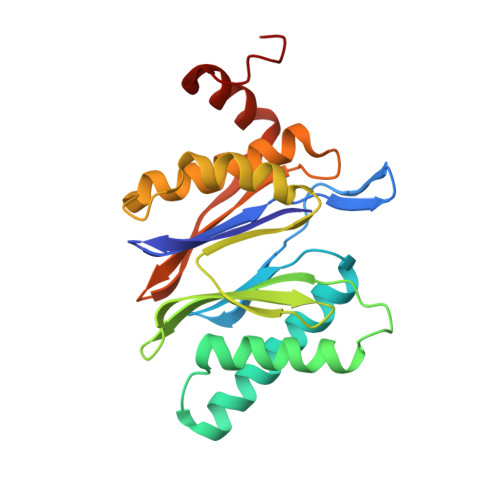
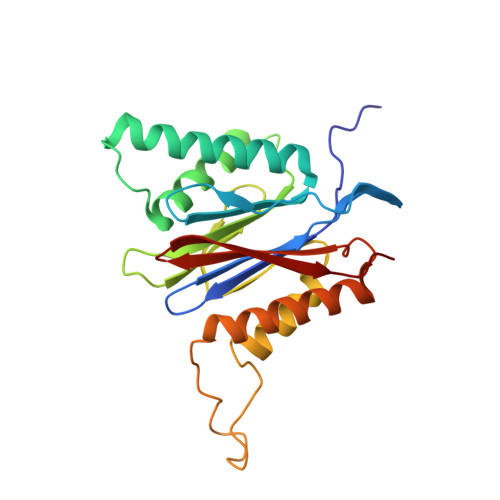
In our screening efforts to identify unique scaffolds from myxobacteria for the drug discovery process, we used LC-SPE-NMR-MS techniques to isolate six linear peptides, termed macyranone A-F, from Cystobacter fuscus MCy9118. The macyranones are characterized by a rare 2-methylmalonamide moiety and an α-amino ketone fragment including an α',β'-epoxyketone in macyranone A. Gene disruption experiments confirmed the biosynthetic gene cluster of the macyranones as PKS/NRPS hybrid. Detailed in silico and phylogenetic analysis unraveled that the biosynthesis involves two conspicuous amide bond formations accomplished by an amidotransferase and a unique condensation domain. The gene cluster provides further insights into the formation of the powerful epoxyketone residue involving an acyl-CoA dehydrogenase and an unconventional free-standing thioesterase. Macyranone A was found to inhibit the chymotrypsin-like activity of the yeast 20S proteasome with an IC50 of 5.9 nM and the human constitutive proteasome and immunoproteasome with IC50 values of 21 and 15 nM, respectively. The β5 subunit of the 20S proteasome was characterized as target by X-ray crystallography revealing an irreversible binding mode similar to the natural product epoxomicin. The presence of the methylmalonamide residue facilitates the stabilization of macyranone A with the active β5 subunit of the proteasome. Macyranone A exhibits a potent inhibitory effect against the parasites Trypanosoma brucei rhodesiense and Leishmania donovani with IC50 values of 1.55 and 0.22 μM, respectively.
Organizational Affiliation:
†Department of Microbial Natural Products, Helmholtz-Institute for Pharmaceutical Research Saarland (HIPS), Helmholtz Centre for Infection Research (HZI) and Pharmaceutical Biotechnology, Saarland University, Campus C2 3, 66123 Saarbrücken, Germany.