Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody.
Apgar, J.R., Mader, M., Agostinelli, R., Benard, S., Bialek, P., Johnson, M., Gao, Y., Krebs, M., Owens, J., Parris, K., St Andre, M., Svenson, K., Morris, C., Tchistiakova, L.(2016) MAbs 8: 1302-1318
- PubMed: 27625211
- DOI: https://doi.org/10.1080/19420862.2016.1215786
- Primary Citation of Related Structures:
5F3B, 5F3H - PubMed Abstract:
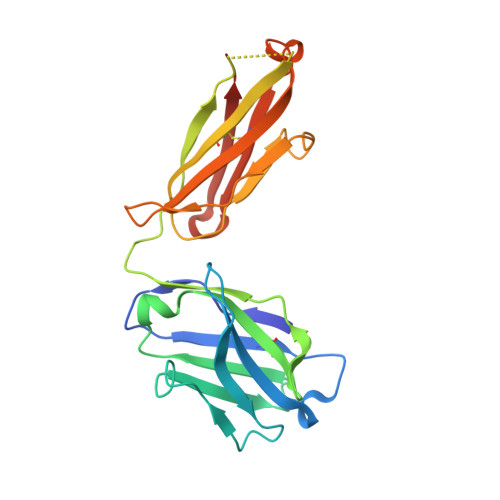
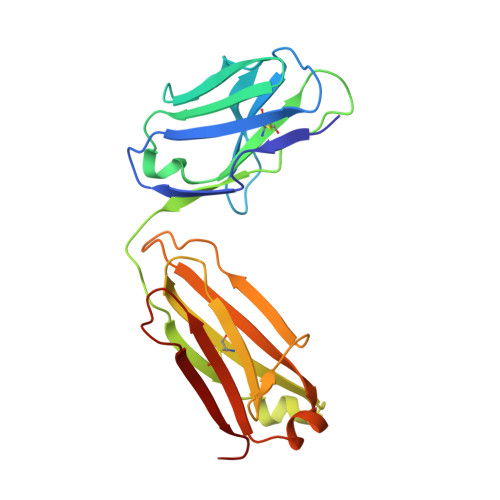
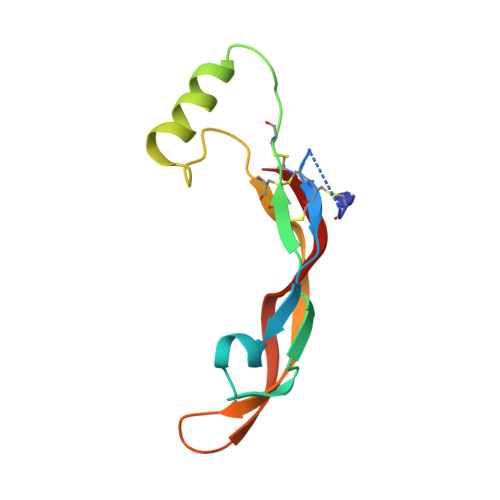
Antibodies are an important class of biotherapeutics that offer specificity to their antigen, long half-life, effector function interaction and good manufacturability. The immunogenicity of non-human-derived antibodies, which can be a major limitation to development, has been partially overcome by humanization through complementarity-determining region (CDR) grafting onto human acceptor frameworks. The retention of foreign content in the CDR regions, however, is still a potential immunogenic liability. Here, we describe the humanization of an anti-myostatin antibody utilizing a 2-step process of traditional CDR-grafting onto a human acceptor framework, followed by a structure-guided approach to further reduce the murine content of CDR-grafted antibodies. To accomplish this, we solved the co-crystal structures of myostatin with the chimeric (Protein Databank (PDB) id 5F3B) and CDR-grafted anti-myostatin antibody (PDB id 5F3H), allowing us to computationally predict the structurally important CDR residues as well as those making significant contacts with the antigen. Structure-based rational design enabled further germlining of the CDR-grafted antibody, reducing the murine content of the antibody without affecting antigen binding. The overall "humanness" was increased for both the light and heavy chain variable regions.
Organizational Affiliation:
a Biomedicine Design, Pfizer Inc. , Cambridge , MA , USA.