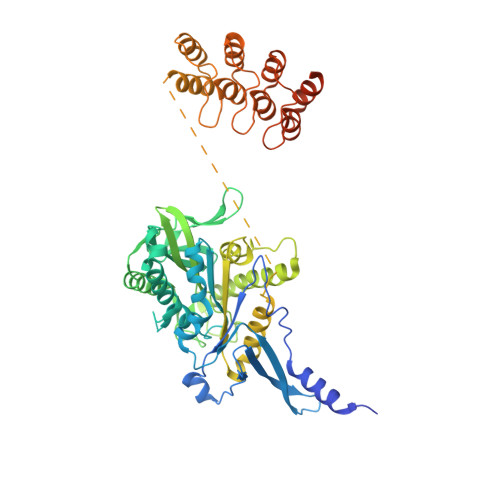
Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin.
Wang, W., Cantos-Fernandes, S., Lv, Y., Kuerban, H., Ahmad, S., Wang, C., Gigant, B.(2017) Nat Commun 8: 70-70
- PubMed: 28694425
- DOI: https://doi.org/10.1038/s41467-017-00091-9
- Primary Citation of Related Structures:
5MIO - PubMed Abstract:
Kinesin-13s are critical microtubule regulators which induce microtubule disassembly in an ATP dependent manner. To clarify their mechanism, we report here the crystal structure of a functional construct of the kinesin-13 Kif2C/MCAK in an ATP-like state and bound to the αβ-tubulin heterodimer, a complex mimicking the species that dissociates from microtubule ends during catalytic disassembly. Our results picture how Kif2C stabilizes a curved tubulin conformation. The Kif2C α4-L12-α5 region undergoes a remarkable 25° rotation upon tubulin binding to target the αβ-tubulin hinge. This movement leads the β5a-β5b motif to interact with the distal end of β-tubulin, whereas the neck and the KVD motif, two specific elements of kinesin-13s, target the α-tubulin distal end. Taken together with the study of Kif2C mutants, our data suggest that stabilization of a curved tubulin is an important contribution to the Kif2C mechanism.Kinesin-13s are microtubule depolymerizing enzymes. Here the authors present the crystal structure of a DARPin fused construct comprising the short neck region and motor domain of kinesin-13 in complex with an αβ-tubulin heterodimer, which shows that kinesin-13 functions by stabilizing a curved tubulin conformation.
Organizational Affiliation:
Department of Central Laboratory, Shanghai Tenth People's Hospital of Tongji University, School of Life Sciences and Technology, Tongji University, Shanghai, 200092, China.