In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus.
Dai, X., Li, Z., Lai, M., Shu, S., Du, Y., Zhou, Z.H., Sun, R.(2017) Nature 541: 112-116
- PubMed: 27992877
- DOI: https://doi.org/10.1038/nature20589
- Primary Citation of Related Structures:
5TC1 - PubMed Abstract:
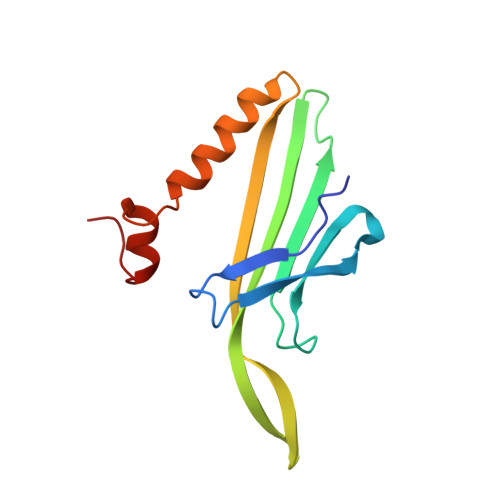
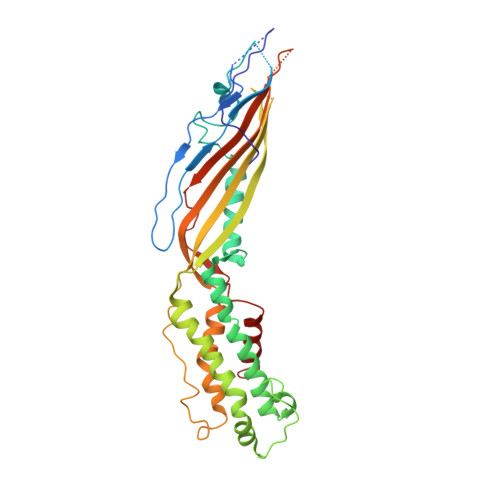
Packaging of the genome into a protein capsid and its subsequent delivery into a host cell are two fundamental processes in the life cycle of a virus. Unlike double-stranded DNA viruses, which pump their genome into a preformed capsid, single-stranded RNA (ssRNA) viruses, such as bacteriophage MS2, co-assemble their capsid with the genome; however, the structural basis of this co-assembly is poorly understood. MS2 infects Escherichia coli via the host 'sex pilus' (F-pilus); it was the first fully sequenced organism and is a model system for studies of translational gene regulation, RNA-protein interactions, and RNA virus assembly. Its positive-sense ssRNA genome of 3,569 bases is enclosed in a capsid with one maturation protein monomer and 89 coat protein dimers arranged in a T = 3 icosahedral lattice. The maturation protein is responsible for attaching the virus to an F-pilus and delivering the viral genome into the host during infection, but how the genome is organized and delivered is not known. Here we describe the MS2 structure at 3.6 Å resolution, determined by electron-counting cryo-electron microscopy (cryoEM) and asymmetric reconstruction. We traced approximately 80% of the backbone of the viral genome, built atomic models for 16 RNA stem-loops, and identified three conserved motifs of RNA-coat protein interactions among 15 of these stem-loops with diverse sequences. The stem-loop at the 3' end of the genome interacts extensively with the maturation protein, which, with just a six-helix bundle and a six-stranded β-sheet, forms a genome-delivery apparatus and joins 89 coat protein dimers to form a capsid. This atomic description of genome-capsid interactions in a spherical ssRNA virus provides insight into genome delivery via the host sex pilus and mechanisms underlying ssRNA-capsid co-assembly, and inspires speculation about the links between nucleoprotein complexes and the origins of viruses.
Organizational Affiliation:
Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), Los Angeles, California 90095, USA.