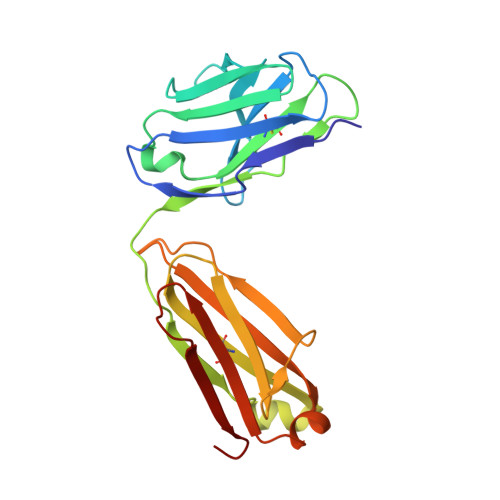
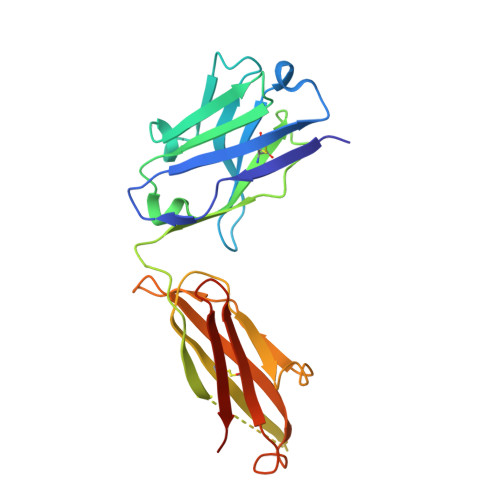
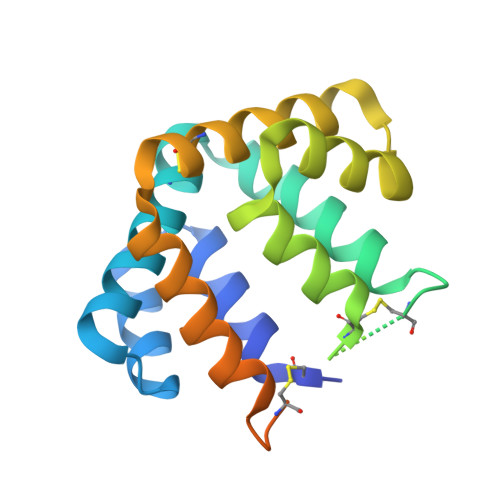
Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement.
Orengo, J.M., Radin, A.R., Kamat, V., Badithe, A., Ben, L.H., Bennett, B.L., Zhong, S., Birchard, D., Limnander, A., Rafique, A., Bautista, J., Kostic, A., Newell, D., Duan, X., Franklin, M.C., Olson, W., Huang, T., Gandhi, N.A., Lipsich, L., Stahl, N., Papadopoulos, N.J., Murphy, A.J., Yancopoulos, G.D.(2018) Nat Commun 9: 1421-1421
- PubMed: 29650949
- DOI: https://doi.org/10.1038/s41467-018-03636-8
- Primary Citation of Related Structures:
5VYF - PubMed Abstract:
Acute allergic symptoms are caused by allergen-induced crosslinking of allergen-specific immunoglobulin E (IgE) bound to Fc-epsilon receptors on effector cells. Desensitization with allergen-specific immunotherapy (SIT) has been used for over a century, but the dominant protective mechanism remains unclear. One consistent observation is increased allergen-specific IgG, thought to competitively block allergen binding to IgE. Here we show that the blocking potency of the IgG response to Cat-SIT is heterogeneous. Next, using two potent, pre-selected allergen-blocking monoclonal IgG antibodies against the immunodominant cat allergen Fel d 1, we demonstrate that increasing the IgG/IgE ratio reduces the allergic response in mice and in cat-allergic patients: a single dose of blocking IgG reduces clinical symptoms in response to nasal provocation (ANCOVA, p = 0.0003), with a magnitude observed at day 8 similar to that reported with years of conventional SIT. This study suggests that simply augmenting the blocking IgG/IgE ratio may reverse allergy.
Organizational Affiliation:
Regeneron Pharmaceuticals Inc., 777 Old Saw Mill River Road, Tarrytown, NY, 10591, USA. jamie.orengo@regeneron.com.