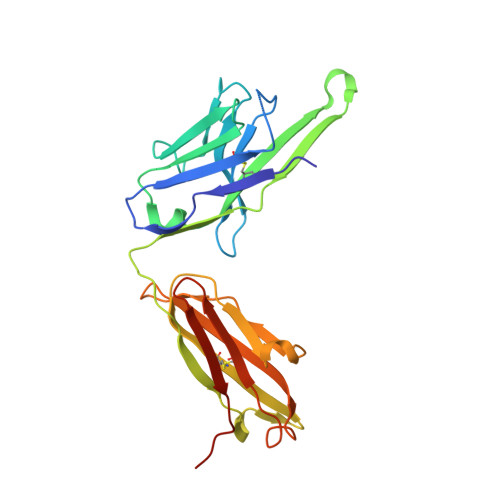
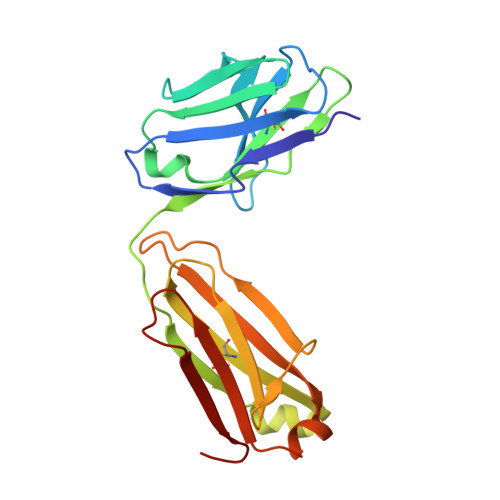
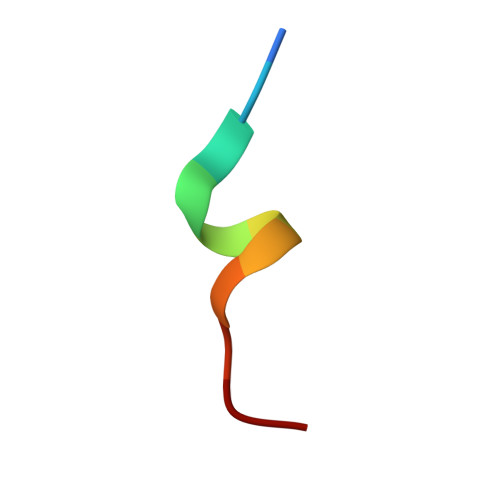
Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Fedechkin, S.O., George, N.L., Wolff, J.T., Kauvar, L.M., DuBois, R.M.(2018) Sci Immunol 3
- PubMed: 29523582
- DOI: https://doi.org/10.1126/sciimmunol.aar3534
- Primary Citation of Related Structures:
5WN9, 5WNA, 5WNB - PubMed Abstract:
Respiratory syncytial virus (RSV) is a top cause of severe lower respiratory tract disease and mortality in young children and the elderly. The viral envelope G glycoprotein contributes to pathogenesis through its roles in host cell attachment and modulation of host immunity. Although the G glycoprotein is a target of protective RSV-neutralizing antibodies, its development as a vaccine antigen has been hindered by its heterogeneous glycosylation and sequence variability outside a conserved central domain (CCD). We describe the cocrystal structures of two high-affinity broadly neutralizing human monoclonal antibodies bound to the RSV G CCD. The antibodies bind to neighboring conformational epitopes, which we named antigenic sites γ1 and γ2, that span a highly conserved surface, illuminating an important region of vulnerability. We further show that isolated RSV G CCD activates the chemokine receptor CX3CR1 and that antibodies block this activity. These studies provide a template for rational vaccine design targeting this key contributor to RSV disease.
Organizational Affiliation:
Department of Biomolecular Engineering, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.