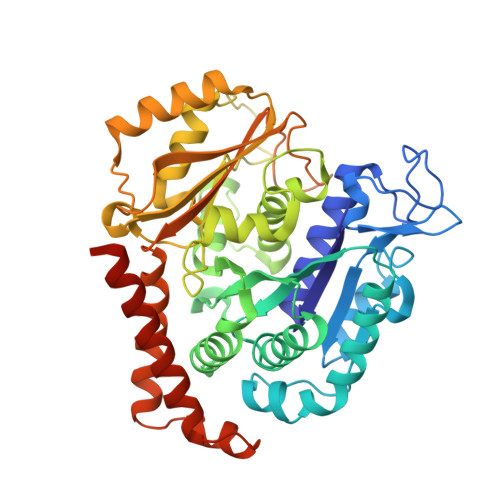
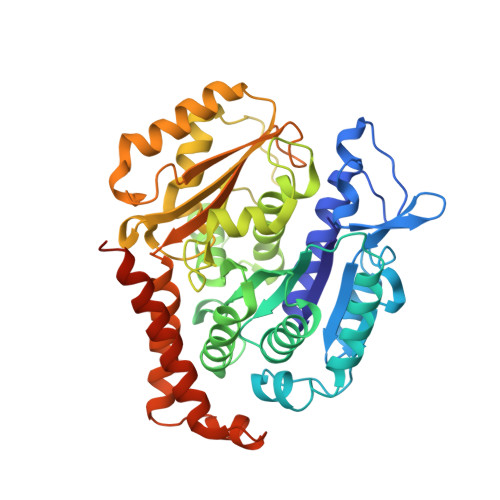
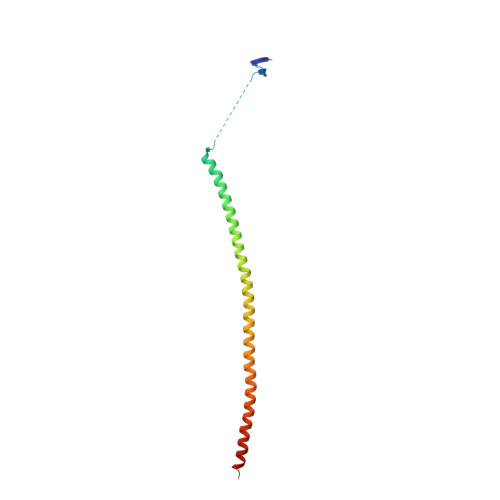
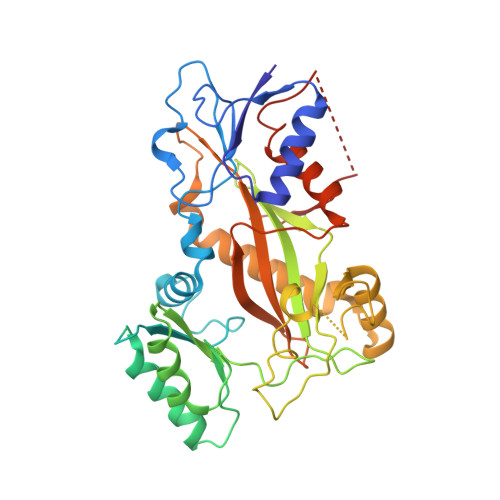
Structure of a benzylidene derivative of 9(10H)-anthracenone in complex with tubulin provides a rationale for drug design.
Cheng, J., Wu, Y., Wang, Y., Wang, C., Wang, Y., Wu, C., Zeng, S., Yu, Y., Chen, Q.(2018) Biochem Biophys Res Commun 495: 185-188
- PubMed: 29102632
- DOI: https://doi.org/10.1016/j.bbrc.2017.10.104
- Primary Citation of Related Structures:
5XLZ - PubMed Abstract:
Microtubules are composed of αβ-tubulin heterodimers and have been treated as highly attractive targets for antitumor drugs. A broad range of agents bind to tubulin and interfere with microtubule assembly, including colchicine binding site inhibitors (CBSIs). Tubulin Polymerization Inhibitor I (TPI1), a benzylidene derivative of 9(10H)-anthracenone, is a CBSI that inhibits the assembly of microtubules. However, for a long time, the design and development of anthracenone family drugs have been hindered by the lack of structural information of the tubulin-agent complex. Here we report a 2.3 Å crystal structure of tubulin complexed with TPI1, the first structure of anthracenone family agents. This complex structure reveals the interactions between TPI1 and tubulin, and thus provides insights into the development of new anthracenone derivatives targeting the colchicine binding site.
Organizational Affiliation:
State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Collaborative Innovation Center of Biotherapy, Chengdu 610041, People's Republic of China.