A Novel Microtubule Inhibitor Overcomes Multidrug Resistance in Tumors.
Ning, N., Yu, Y., Wu, M., Zhang, R., Zhang, T., Zhu, C., Huang, L., Yun, C.H., Benes, C.H., Zhang, J., Deng, X., Chen, Q., Ren, R.(2018) Cancer Res 78: 5949-5957
- PubMed: 30135190
- DOI: https://doi.org/10.1158/0008-5472.CAN-18-0455
- Primary Citation of Related Structures:
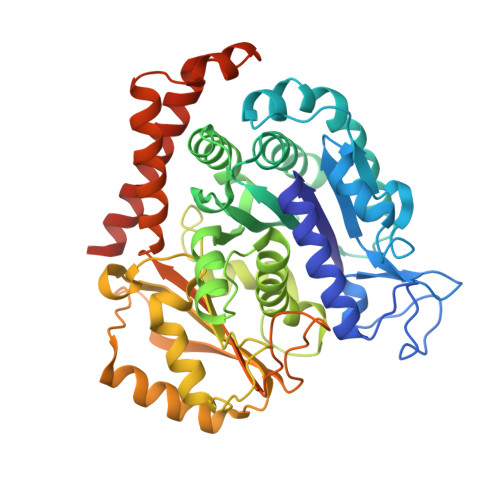
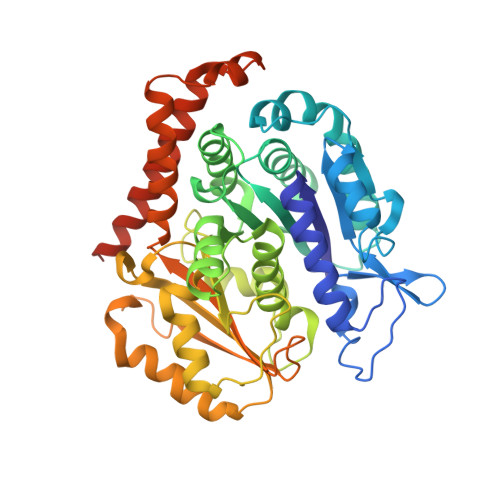
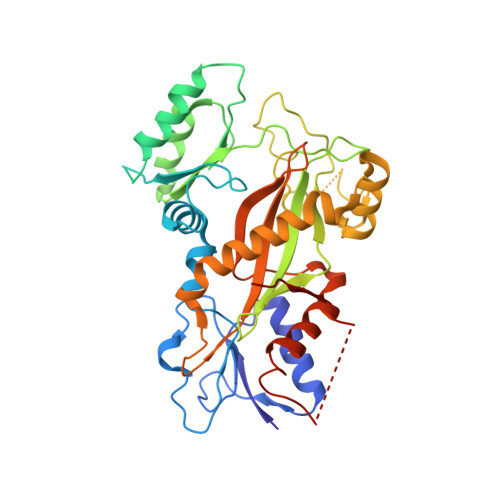
5YZ3 - PubMed Abstract:
Microtubule inhibitors as chemotherapeutic drugs are widely used for cancer treatment. However, the development of multidrug resistance (MDR) in cancer is a major challenge for microtubule inhibitors in their clinical implementation. From a high-throughput drug screen using cells transformed by oncogenic RAS, we identify a lead heteroaryl amide compound that blocks cell proliferation. Analysis of the structure-activity relationship indicated that this series of scaffolds (exemplified by MP-HJ-1b) represents a potent inhibitor of tumor cell growth. MP-HJ-1b showed activities against a panel of more than 1,000 human cancer cell lines with a wide variety of tissue origins. This compound depolymerized microtubules and affected spindle formation. It also induced the spike-like conformation of microtubules in vitro and in vivo , which is different from typical microtubule modulators. Structural analysis revealed that this series of compounds bound the colchicine pocket at the intra-dimer interface, although mostly not overlapping with colchicine binding. MP-HJ-1b displayed favorable pharmacological properties for overcoming tumor MDR, both in vitro and in vivo Taken together, our data reveal a novel scaffold represented by MP-HJ-1b that can be developed as a cancer therapeutic against tumors with MDR. Significance: Paclitaxel is a widely used chemotherapeutic drug in patients with multiple types of cancer. However, resistance to paclitaxel is a challenge. This study describes a novel class of microtubule inhibitors with the ability to circumvent multidrug resistance across multiple tumor cell lines. Cancer Res; 78(20); 5949-57. ©2018 AACR .
Organizational Affiliation:
State Key Laboratory for Medical Genomics, Shanghai Institute of Hematology, Collaborative Innovation Center of Hematology, Collaborative Innovation Center of System Biology, Ruijin Hospital, School of Life Sciences and Biotechnology and School of Medicine, Shanghai Jiao Tong University, Shanghai, China.