Discovery of DS-6930, a potent selective PPAR gamma modulator. Part II: Lead optimization.
Shinozuka, T., Tsukada, T., Fujii, K., Tokumaru, E., Shimada, K., Onishi, Y., Matsui, Y., Wakimoto, S., Kuroha, M., Ogata, T., Araki, K., Ohsumi, J., Sawamura, R., Watanabe, N., Yamamoto, H., Fujimoto, K., Tani, Y., Mori, M., Tanaka, J.(2018) Bioorg Med Chem 26: 5099-5117
- PubMed: 30220602
- DOI: https://doi.org/10.1016/j.bmc.2018.09.005
- Primary Citation of Related Structures:
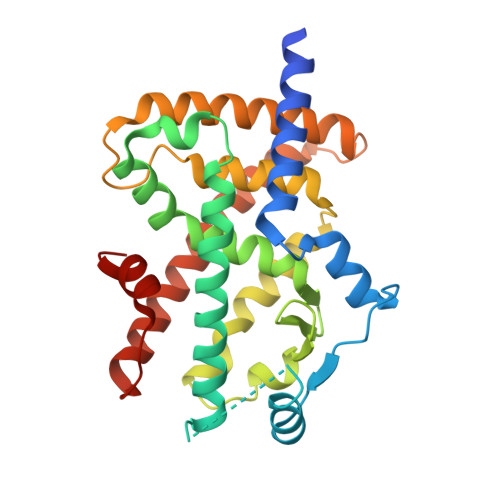
5Z6S - PubMed Abstract:
Attempts were made to reduce the lipophilicity of previously synthesized compound (II) for the avoidance of hepatotoxicity. The replacement of the left-hand side benzene with 2-pyridine resulted in the substantial loss of potency. Because poor membrane permeability was responsible for poor potency in vitro, the adjustment of lipophilicity was examined, which resulted in the discovery of dimethyl pyridine derivative (I, DS-6930). In preclinical studies, DS-6930 demonstrated high PPARγ agonist potency with robust plasma glucose reduction. DS-6930 maintained diminished PPARγ-related adverse effects upon toxicological evaluation in vivo, and demonstrated no hepatotoxicity. Cofactor recruitment assay showed that several cofactors, such as RIP140 and PGC1, were significantly recruited, whereas several canonical factors was not affected. This selective cofactor recruitment was caused due to the distinct binding mode of DS-6930. The calcium salt, DS-6930b, which is expected to be an effective inducer of insulin sensitization without edema, could be evaluated clinically in T2DM patients.
Organizational Affiliation:
R&D Division, Daiichi Sankyo Co., Ltd., 1-2-58 Hiromachi, Shinagawa-ku, Tokyo 140-8710, Japan. Electronic address: sinozu.xf6@gmail.com.