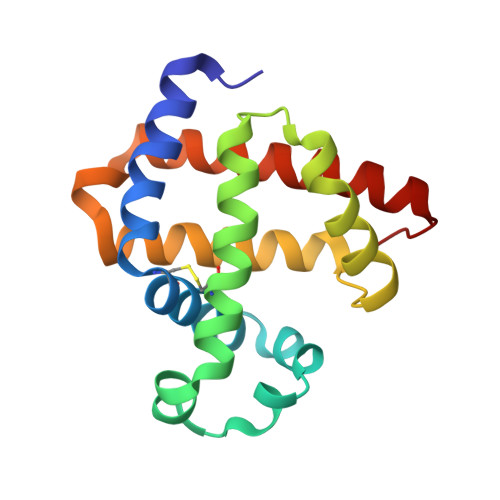
Regulation of both the structure and function by a de novo designed disulfide bond: a case study of heme proteins in myoglobin
Yin, L.L., Yuan, H., Du, K.J., He, B., Gao, S.Q., Wen, G.B., Tan, X., Lin, Y.W.(2018) Chem Commun (Camb) 54: 4356-4359
- PubMed: 29645029
- DOI: https://doi.org/10.1039/c8cc01646a
- Primary Citation of Related Structures:
5ZEO - PubMed Abstract:
A de novo designed intramolecular disulfide bond in myoglobin, resembling that in cytoglobin without structural evidence, was confirmed by an X-ray structure for the first time and was demonstrated to regulate both the structure and function of this protein, which fulfills the design of an artificial dehaloperoxidase, with an activity exceeding that of a native enzyme.
Organizational Affiliation:
School of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China. linlinying@hotmail.com ywlin@usc.edu.cn.