Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion.
Wang, H., Barnes, C.O., Yang, Z., Nussenzweig, M.C., Bjorkman, P.J.(2018) Cell Host Microbe 24: 579-592.e4
- PubMed: 30308160
- DOI: https://doi.org/10.1016/j.chom.2018.09.003
- Primary Citation of Related Structures:
6CM3, 6EDU - PubMed Abstract:
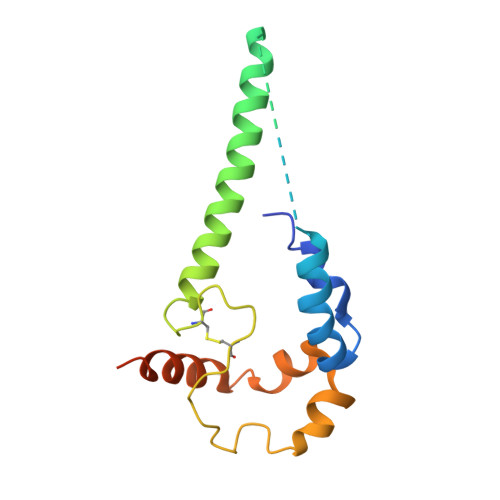
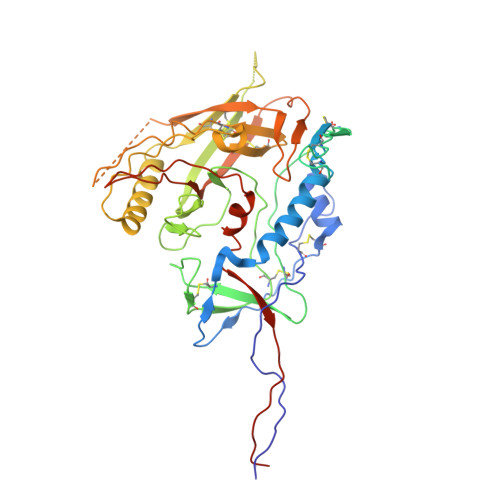
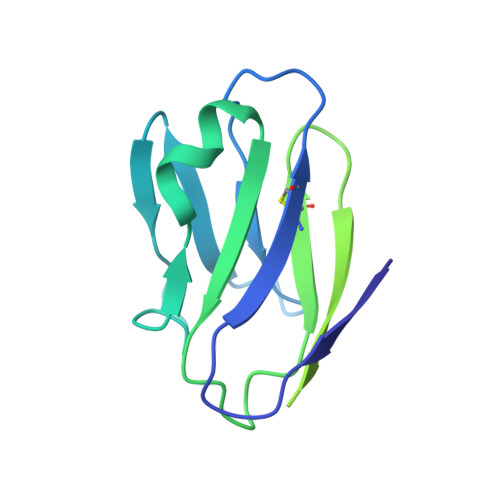
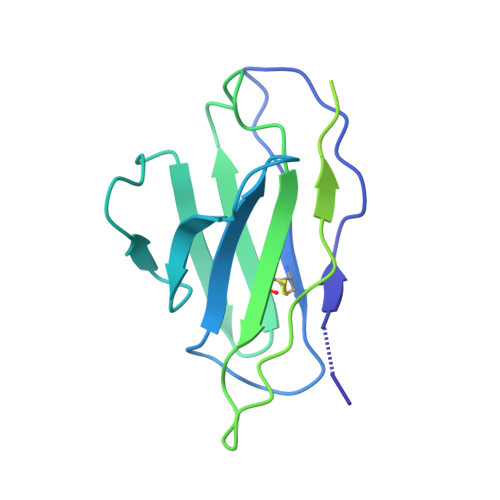
HIV-1 Env, a trimer of gp120-gp41 heterodimers, mediates membrane fusion after binding host receptor CD4. Receptor binding displaces V1V2 loops from Env's apex, allowing coreceptor binding and opening Env to enable gp41-mediated fusion. We present 3.54 Å and 4.06 Å cryoelectron microscopy structures of partially open soluble native-like Env trimers (SOSIPs) bound to CD4. One structure, a complex with a coreceptor-mimicking antibody that binds both CD4 and gp120, stabilizes the displaced V1V2 and reveals its structure. Comparing partially and fully open Envs with closed Envs shows that gp41 rearrangements are independent of the CD4-induced rearrangements that result in V1V2 displacement and formation of a 4-stranded bridging sheet. These findings suggest ordered conformational changes before coreceptor binding: (1) gp120 opening inducing side-chain rearrangements and a compact gp41 central helix conformation, and (2) 4-stranded bridging-sheet formation and V1V2 displacement. These analyses illuminate potential receptor-induced Env changes and inform design of therapeutics disrupting viral entry.
Organizational Affiliation:
Division of Biology and Biological Engineering, California Institute of Technology, 1200 E. California Boulevard, Pasadena, CA 91125, USA.