Structural Modification of the 3,4,5-Trimethoxyphenyl Moiety in the Tubulin Inhibitor VERU-111 Leads to Improved Antiproliferative Activities.
Wang, Q., Arnst, K.E., Wang, Y., Kumar, G., Ma, D., Chen, H., Wu, Z., Yang, J., White, S.W., Miller, D.D., Li, W.(2018) J Med Chem 61: 7877-7891
- PubMed: 30122035
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00827
- Primary Citation of Related Structures:
6D88 - PubMed Abstract:
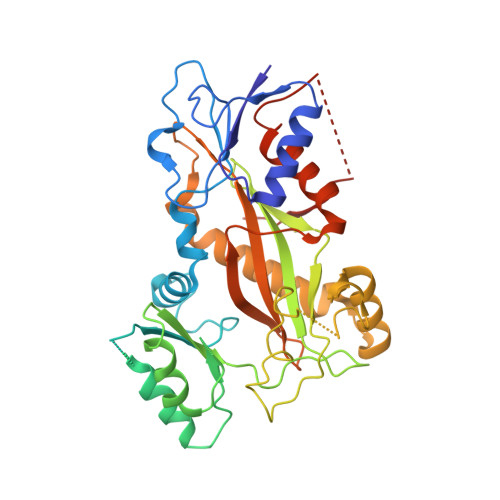
Colchicine binding site inhibitors (CBSIs) hold great potential in developing new generations of antimitotic drugs. Unlike existing tubulin inhibitors such as paclitaxel, they are generally much less susceptible to resistance caused by the overexpression of drug efflux pumps. The 3,4,5-trimethoxyphenyl (TMP) moiety is a critical component present in many CBSIs, playing an important role in maintaining suitable molecular conformations of CBSIs and contributing to their high binding affinities to tubulin. Previously reported modifications to the TMP moiety in a variety of scaffolds of CBSIs have usually resulted in reduced antiproliferative potency. We previously reported a potent CBSI, VERU-111, that also contains the TMP moiety. Herein, we report the discovery of a VERU-111 analogue 13f that is significantly more potent than VERU-111. The X-ray crystal structure of 13f in complex with tubulin confirms its direct binding to the colchicine site. In addition, 13f exhibited a strong inhibitory effect on tumor growth in vivo.
Organizational Affiliation:
Department of Pharmaceutical Sciences, College of Pharmacy , University of Tennessee Health Science Center , Memphis , Tennessee 38163 , United States.