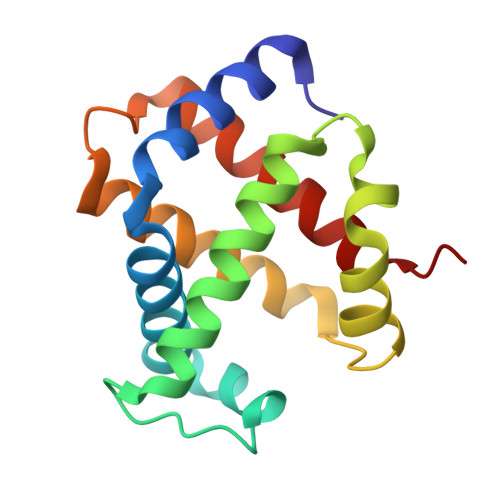
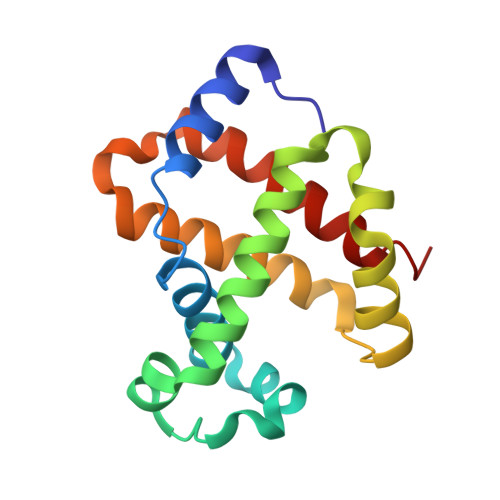
Rational modification of vanillin derivatives to stereospecifically destabilize sickle hemoglobin polymer formation.
Deshpande, T.M., Pagare, P.P., Ghatge, M.S., Chen, Q., Musayev, F.N., Venitz, J., Zhang, Y., Abdulmalik, O., Safo, M.K.(2018) Acta Crystallogr D Struct Biol 74: 956-964
- PubMed: 30289405
- DOI: https://doi.org/10.1107/S2059798318009919
- Primary Citation of Related Structures:
6DI4 - PubMed Abstract:
Increasing the affinity of hemoglobin for oxygen represents a feasible and promising therapeutic approach for sickle cell disease by mitigating the primary pathophysiological event, i.e. the hypoxia-induced polymerization of sickle hemoglobin (Hb S) and the concomitant erythrocyte sickling. Investigations on a novel synthetic antisickling agent, SAJ-310, with improved and sustained antisickling activity have previously been reported. To further enhance the biological effects of SAJ-310, a structure-based approach was employed to modify this compound to specifically inhibit Hb S polymer formation through interactions which perturb the Hb S polymer-stabilizing αF-helix, in addition to primarily increasing the oxygen affinity of hemoglobin. Three compounds, TD-7, TD-8 and TD-9, were synthesized and studied for their interactions with hemoglobin at the atomic level, as well as their functional and antisickling activities in vitro. X-ray crystallographic studies with liganded hemoglobin in complex with TD-7 showed the predicted mode of binding, although the interaction with the αF-helix was not as strong as expected. These findings provide important insights and guidance towards the development of molecules that would be expected to bind and make stronger interactions with the αF-helix, resulting in more efficacious novel therapeutics for sickle cell disease.
Organizational Affiliation:
Department of Medicinal Chemistry, School of Pharmacy, Virginia Commonwealth University, 800 East Leigh Street, Richmond, VA 23219, USA.