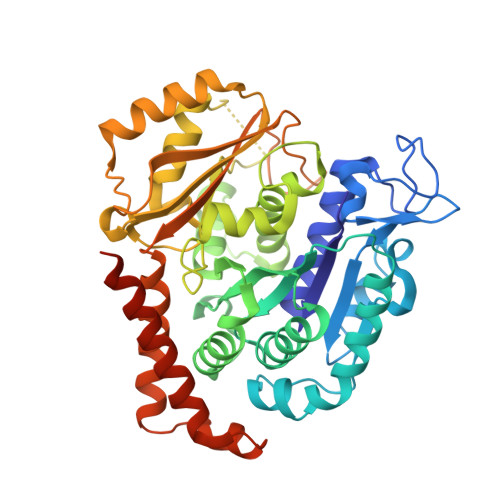
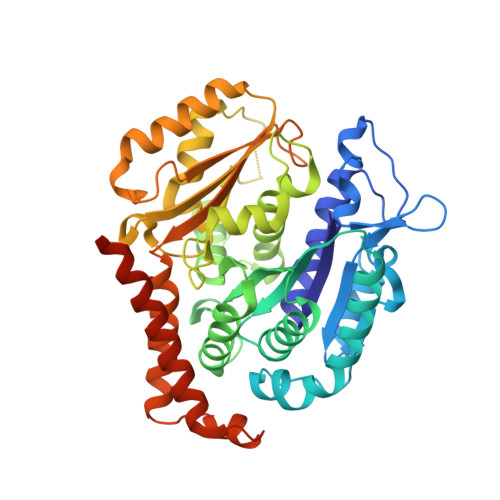
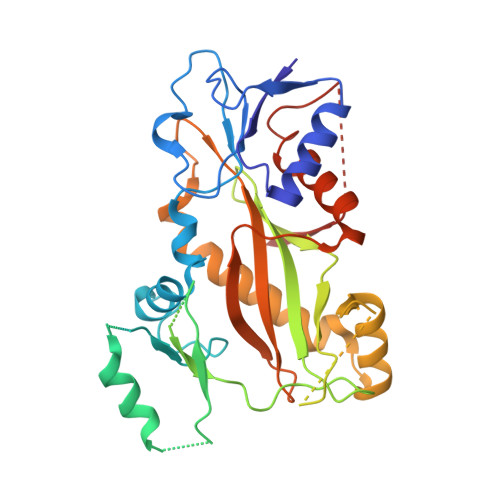
Structural Basis of Colchicine-Site targeting Acylhydrazones active against Multidrug-Resistant Acute Lymphoblastic Leukemia.
Cury, N.M., Muhlethaler, T., Laranjeira, A.B.A., Canevarolo, R.R., Zenatti, P.P., Lucena-Agell, D., Barasoain, I., Song, C., Sun, D., Dovat, S., Yunes, R.A., Prota, A.E., Steinmetz, M.O., Diaz, J.F., Yunes, J.A.(2019) iScience 21: 95-109
- PubMed: 31655259
- DOI: https://doi.org/10.1016/j.isci.2019.10.003
- Primary Citation of Related Structures:
6F7C - PubMed Abstract:
Tubulin is one of the best validated anti-cancer targets, but most anti-tubulin agents have unfavorable therapeutic indexes. Here, we characterized the tubulin-binding activity, the mechanism of action, and the in vivo anti-leukemia efficacy of three 3,4,5-trimethoxy-N-acylhydrazones. We show that all compounds target the colchicine-binding site of tubulin and that none is a substrate of ABC transporters. The crystal structure of the tubulin-bound N-(1'-naphthyl)-3,4,5-trimethoxybenzohydrazide (12) revealed steric hindrance on the T7 loop movement of β-tubulin, thereby rendering tubulin assembly incompetent. Using dose escalation and short-term repeated dose studies, we further report that this compound class is well tolerated to >100 mg/kg in mice. We finally observed that intraperitoneally administered compound 12 significantly prolonged the overall survival of mice transplanted with both sensitive and multidrug-resistant acute lymphoblastic leukemia (ALL) cells. Taken together, this work describes promising colchicine-site-targeting tubulin inhibitors featuring favorable therapeutic effects against ALL and multidrug-resistant cells.
Organizational Affiliation:
Laboratório de Biologia Molecular, Centro Infantil Boldrini, Rua Dr. Gabriel Porto 1270, Campinas 13083-210, Brazil; Graduate Program in Genetics and Molecular Biology, State University of Campinas, Campinas 13083-210, Brazil.