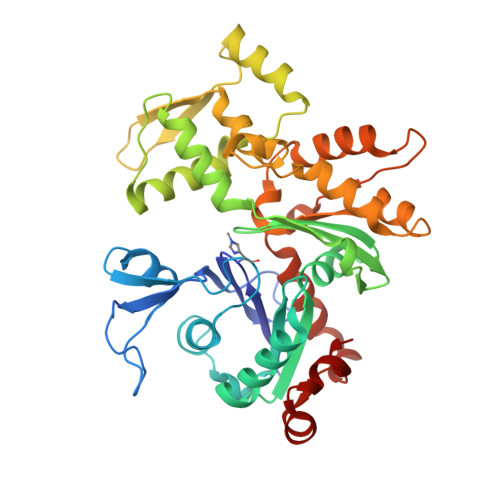
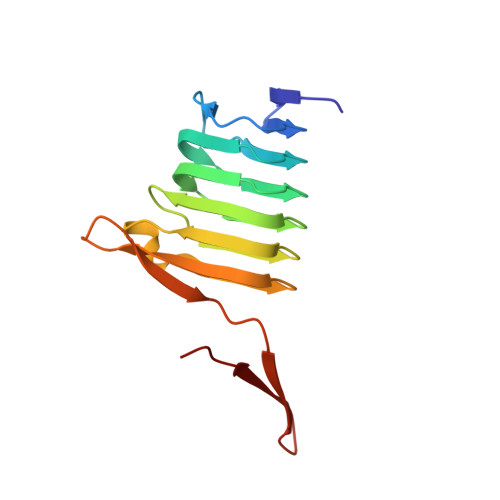
Structural basis of actin monomer re-charging by cyclase-associated protein.
Kotila, T., Kogan, K., Enkavi, G., Guo, S., Vattulainen, I., Goode, B.L., Lappalainen, P.(2018) Nat Commun 9: 1892-1892
- PubMed: 29760438
- DOI: https://doi.org/10.1038/s41467-018-04231-7
- Primary Citation of Related Structures:
6FM2 - PubMed Abstract:
Actin polymerization powers key cellular processes, including motility, morphogenesis, and endocytosis. The actin turnover cycle depends critically on "re-charging" of ADP-actin monomers with ATP, but whether this reaction requires dedicated proteins in cells, and the underlying mechanism, have remained elusive. Here we report that nucleotide exchange catalyzed by the ubiquitous cytoskeletal regulator cyclase-associated protein (CAP) is critical for actin-based processes in vivo. We determine the structure of the CAP-actin complex, which reveals that nucleotide exchange occurs in a compact, sandwich-like complex formed between the dimeric actin-binding domain of CAP and two ADP-actin monomers. In the crystal structure, the C-terminal tail of CAP associates with the nucleotide-sensing region of actin, and this interaction is required for rapid re-charging of actin by both yeast and mammalian CAPs. These data uncover the conserved structural basis and biological role of protein-catalyzed re-charging of actin monomers.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki, 00014, Helsinki, Finland.