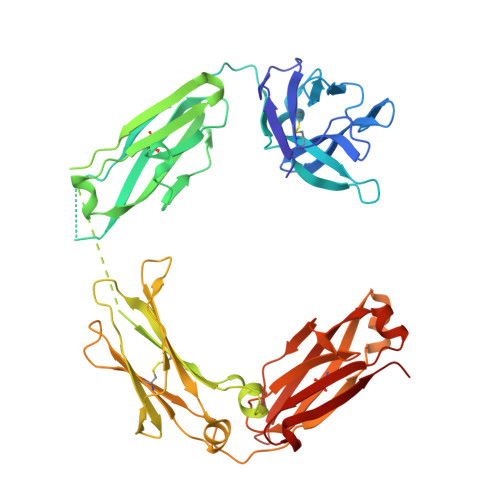
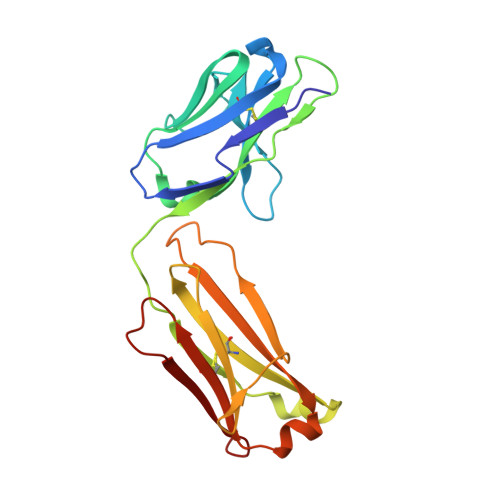
Structure of a Therapeutic Full-Length Anti-NPRA IgG4 Antibody: Dissecting Conformational Diversity.
Blech, M., Horer, S., Kuhn, A.B., Kube, S., Goddeke, H., Kiefer, H., Zang, Y., Alber, Y., Kast, S.M., Westermann, M., Tully, M.D., Schafer, L.V., Garidel, P.(2019) Biophys J 116: 1637-1649
- PubMed: 31023536
- DOI: https://doi.org/10.1016/j.bpj.2019.03.036
- Primary Citation of Related Structures:
6GFE - PubMed Abstract:
We report the x-ray crystal structure of intact, full-length human immunoglobulin (IgG4) at 1.8 Å resolution. The data for IgG4 (S228P), an antibody targeting the natriuretic peptide receptor A, show a previously unrecognized type of Fab-Fc orientation with a distorted λ-shape in which one Fab-arm is oriented toward the Fc portion. Detailed structural analysis by x-ray crystallography and molecular simulations suggest that this is one of several conformations coexisting in a dynamic equilibrium state. These results were confirmed by small angle x-ray scattering in solution. Furthermore, electron microscopy supported these findings by preserving molecule classes of different conformations. This study fosters our understanding of IgG4 in particular and our appreciation of antibody flexibility in general. Moreover, we give insights into potential biological implications, specifically for the interaction of human anti-natriuretic peptide receptor A IgG4 with the neonatal Fc receptor, Fcγ receptors, and complement-activating C1q by considering conformational flexibility.
Organizational Affiliation:
Innovation Unit, Pharmaceutical Development Biologics, Biberach (Riss), Germany. Electronic address: michaela.blech@boehringer-ingelheim.com.