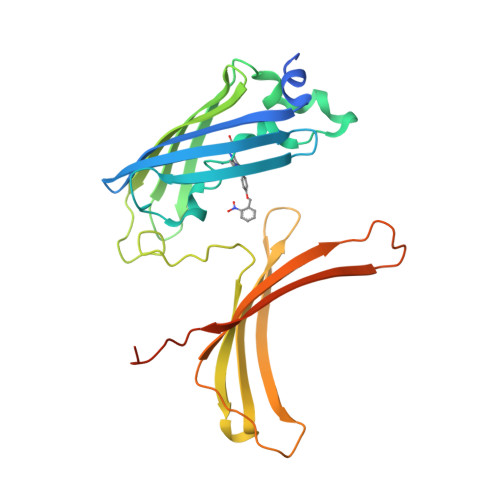
Crystal structure of a domain-swapped photoactivatable sfGFP variant provides evidence for GFP folding pathway.
Kesgin-Schaefer, S., Heidemann, J., Puchert, A., Koelbel, K., Yorke, B.A., Huse, N., Pearson, A.R., Uetrecht, C., Tidow, H.(2019) FEBS J 286: 2329-2340
- PubMed: 30817081
- DOI: https://doi.org/10.1111/febs.14797
- Primary Citation of Related Structures:
6H01 - PubMed Abstract:
Photoactivatable fluorescent proteins (PA-FPs) are a powerful non-invasive tool in high-resolution live-cell imaging. They can be converted from an inactive to an active form by light, enabling the spatial and temporal trafficking of proteins and cell dynamics. PA-FPs have been previously generated by mutating selected residues in the chromophore or in its close proximity. A new strategy to generate PA-FPs is the genetic incorporation of unnatural amino acids (UAAs) containing photocaged groups using unique suppressor tRNA/aminoacyl-tRNA synthetase pairs. We set out to develop a photoactivatable GFP variant suitable for time-resolved structural studies. Here, we report the crystal structure of superfolder GFP (sfGFP) containing the UAA ortho-nitrobenzyl-tyrosine (ONBY) at position 66 and its spectroscopic characterization. Surprisingly, the crystal structure (to 2.7 Å resolution) reveals a dimeric domain-swapped arrangement of sfGFP66ONBY with residues 1-142 of one molecule associating with residues 148-234 from another molecule. This unusual domain-swapped structure supports a previously postulated GFP folding pathway that proceeds via an equilibrium intermediate.
Organizational Affiliation:
The Hamburg Centre for Ultrafast Imaging, Germany.