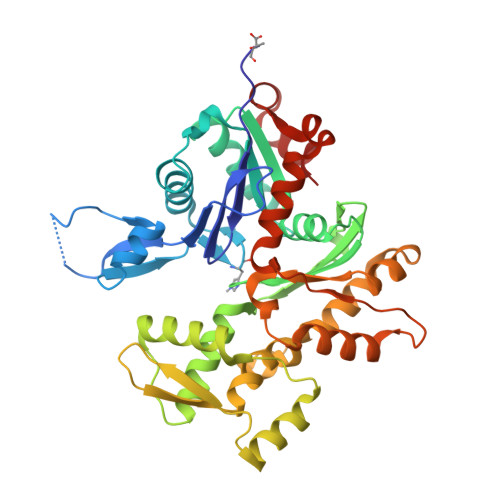
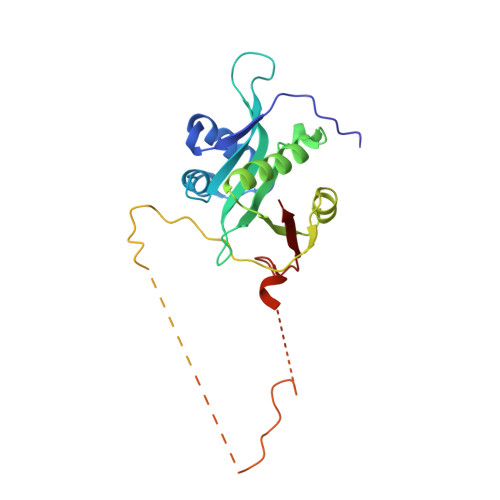
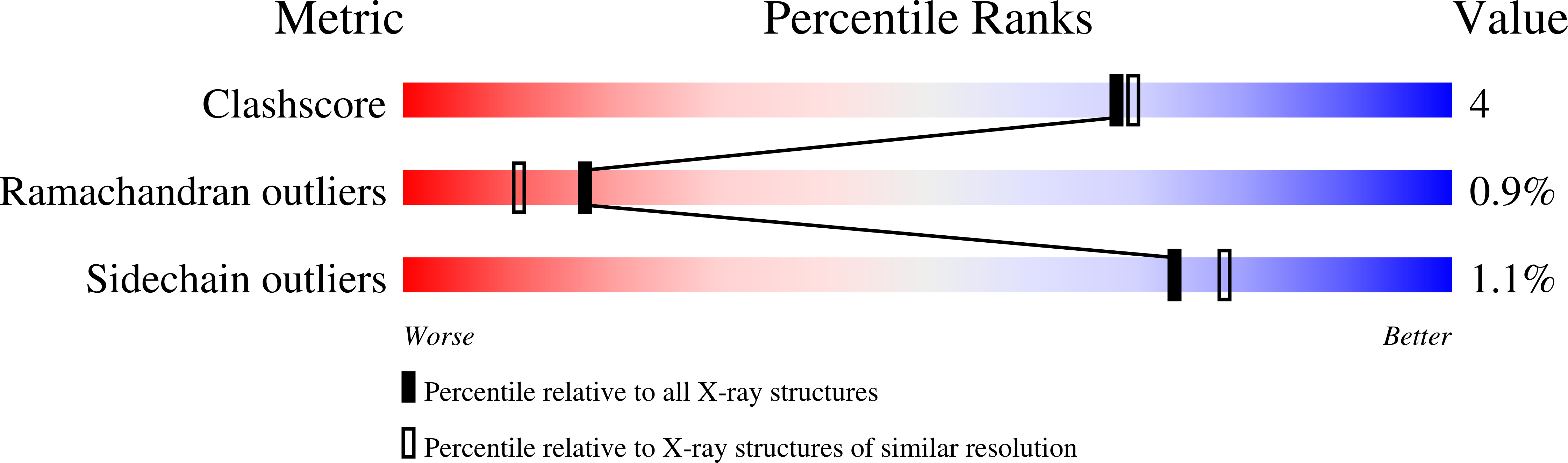Mechanism of actin N-terminal acetylation.
Rebowski, G., Boczkowska, M., Drazic, A., Ree, R., Goris, M., Arnesen, T., Dominguez, R.(2020) Sci Adv 6: eaay8793-eaay8793
- PubMed: 32284999
- DOI: https://doi.org/10.1126/sciadv.aay8793
- Primary Citation of Related Structures:
6NAS, 6NBE, 6NBW - PubMed Abstract:
About 80% of human proteins are amino-terminally acetylated (Nt-acetylated) by one of seven Nt-acetyltransferases (NATs). Actin, the most abundant protein in the cytoplasm, has its own dedicated NAT, NAA80, which acts posttranslationally and affects cytoskeleton assembly and cell motility. Here, we show that NAA80 does not associate with filamentous actin in cells, and its natural substrate is the monomeric actin-profilin complex, consistent with Nt-acetylation preceding polymerization. NAA80 Nt-acetylates actin-profilin much more efficiently than actin alone, suggesting that profilin acts as a chaperone for actin Nt-acetylation. We determined crystal structures of the NAA80-actin-profilin ternary complex, representing different actin isoforms and different states of the catalytic reaction and revealing the first structure of NAT-substrate complex at atomic resolution. The structural, biochemical, and cellular analysis of mutants shows how NAA80 has evolved to specifically recognize actin among all cellular proteins while targeting all six actin isoforms, which differ the most at the amino terminus.
Organizational Affiliation:
Department of Physiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.