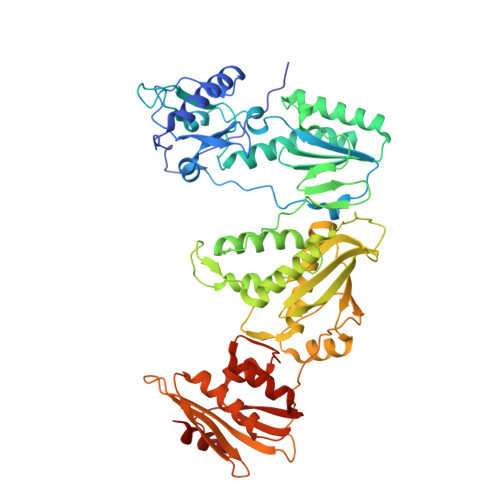
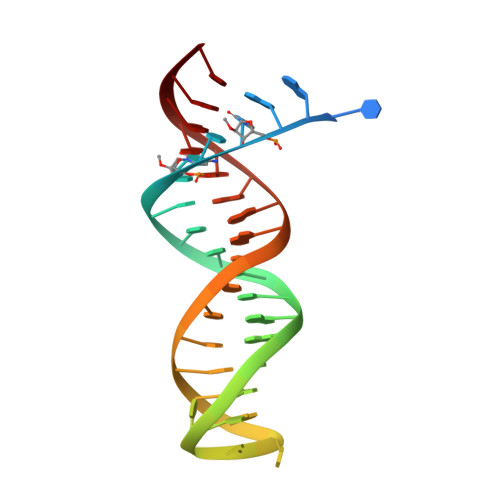
Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1.
Ruiz, F.X., Hoang, A., Das, K., Arnold, E.(2019) J Med Chem 62: 9996-10002
- PubMed: 31603676
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01289
- Primary Citation of Related Structures:
6O9E - PubMed Abstract:
HIV-1 reverse transcriptase (RT) is an essential enzyme, targeting half of approved anti-AIDS drugs. While nucleoside RT inhibitors (NRTIs) are DNA chain terminators, the nucleotide-competing RT inhibitor (NcRTI) INDOPY-1 blocks dNTP binding to RT. Lack of structural information hindered INDOPY-1 improvement. Here we report the HIV-1 RT/DNA/INDOPY-1 crystal structure, revealing a unique mode of inhibitor binding at the polymerase active site without involving catalytic metal ions. The structure may enable new strategies for developing NcRTIs.
Organizational Affiliation:
Rega Institute for Medical Research , 3000 Leuven , Belgium.