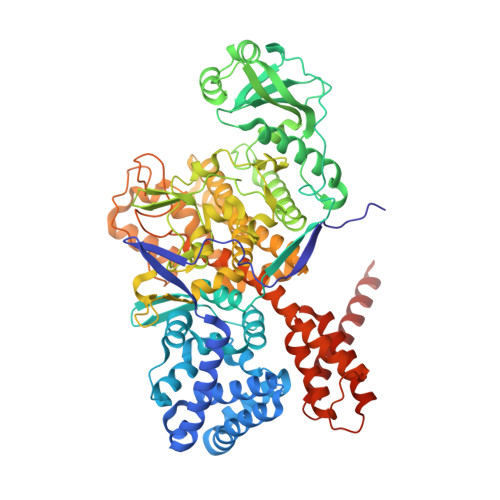
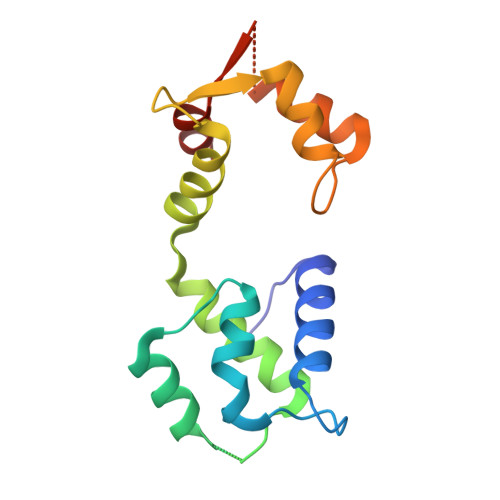
Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ.
Sulpizio, A., Minelli, M.E., Wan, M., Burrowes, P.D., Wu, X., Sanford, E.J., Shin, J.H., Williams, B.C., Goldberg, M.L., Smolka, M.B., Mao, Y.(2019) Elife 8
- PubMed: 31682223
- DOI: https://doi.org/10.7554/eLife.51162
- Primary Citation of Related Structures:
6PLM - PubMed Abstract:
Pseudokinases are considered to be the inactive counterparts of conventional protein kinases and comprise approximately 10% of the human and mouse kinomes. Here, we report the crystal structure of the Legionella pneumophila effector protein, SidJ, in complex with the eukaryotic Ca 2+ -binding regulator, calmodulin (CaM). The structure reveals that SidJ contains a protein kinase-like fold domain, which retains a majority of the characteristic kinase catalytic motifs. However, SidJ fails to demonstrate kinase activity. Instead, mass spectrometry and in vitro biochemical analyses demonstrate that SidJ modifies another Legionella effector SdeA, an unconventional phosphoribosyl ubiquitin ligase, by adding glutamate molecules to a specific residue of SdeA in a CaM-dependent manner. Furthermore, we show that SidJ-mediated polyglutamylation suppresses the ADP-ribosylation activity. Our work further implies that some pseudokinases may possess ATP-dependent activities other than conventional phosphorylation.
Organizational Affiliation:
Weill Institute for Cell and Molecular Biology, Cornell University, Ithaca, United States.