Structure, stability and aggregation propensity of a Ribonuclease A-Onconase chimera.
Esposito, L., Donnarumma, F., Ruggiero, A., Leone, S., Vitagliano, L., Picone, D.(2019) Int J Biol Macromol 133: 1125-1133
- PubMed: 31026530
- DOI: https://doi.org/10.1016/j.ijbiomac.2019.04.164
- Primary Citation of Related Structures:
6QMN - PubMed Abstract:
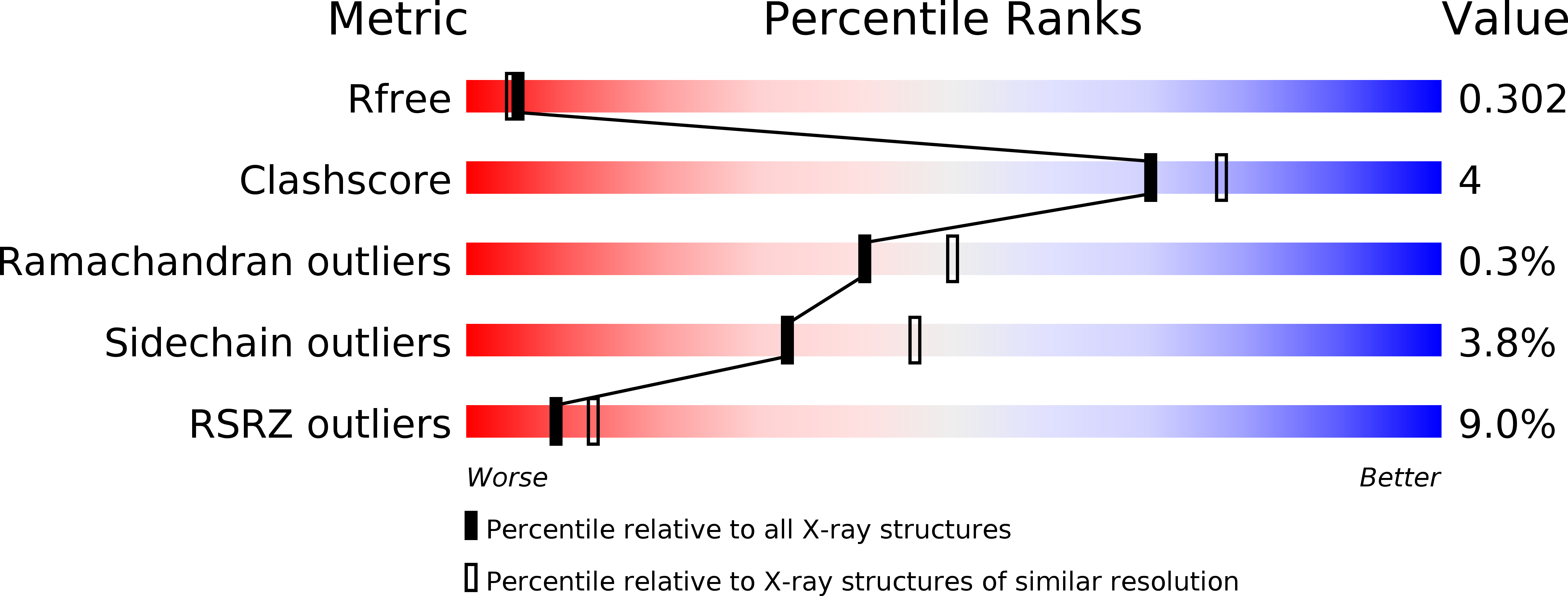
Structural roles of loop regions are frequently overlooked in proteins. Nevertheless, they may be key players in the definition of protein topology and in the self-assembly processes occurring through domain swapping. We here investigate the effects on structure and stability of replacing the loop connecting the last two β-strands of RNase A with the corresponding region of the more thermostable Onconase. The crystal structure of this chimeric variant (RNaseA-ONC) shows that its terminal loop size better adheres to the topological rules for the design of stabilized proteins, proposed by Baker and coworkers [43]. Indeed, RNaseA-ONC displays a thermal stability close to that of RNase A, despite the lack of Pro at position 114, which, due to its propensity to favor a cis peptide bond, has been identified as an important stabilizing factor of the native protein. Accordingly, RNaseA-ONC is significantly more stable than RNase A variants lacking Pro114; RNaseA-ONC also displays a higher propensity to form oligomers in native conditions when compared to either RNase A or Onconase. This finding demonstrates that modifications of terminal loops should to be carefully controlled in terms of size and sequence to avoid unwanted and/or potentially harmful aggregation processes.
Organizational Affiliation:
CNR Istituto di Biostrutture e Bioimmagini, Via Mezzocannone 16, I-80134 Napoli, Italy. Electronic address: luciana.esposito@cnr.it.