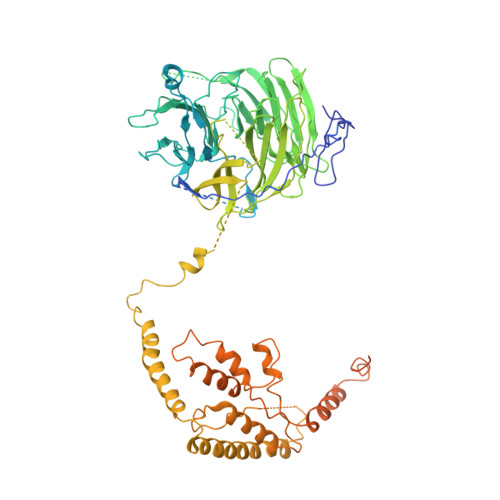
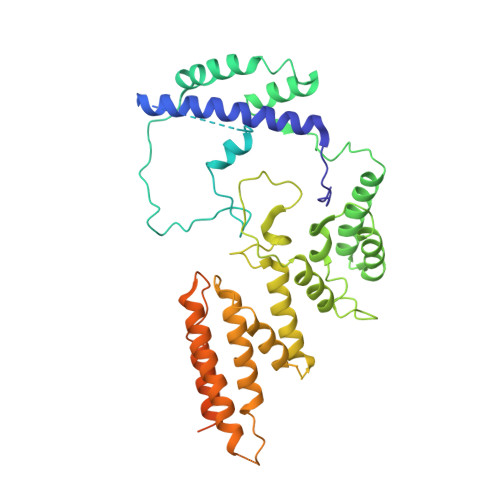
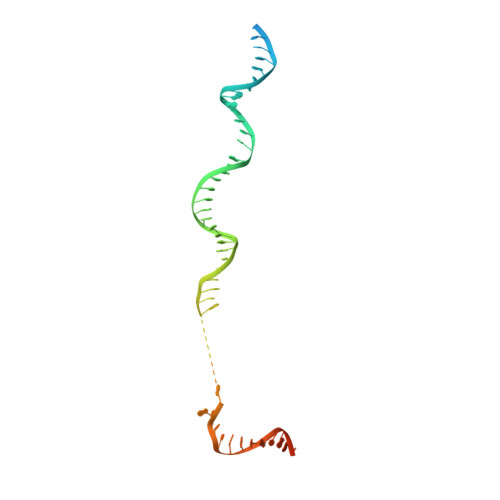
Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Sadian, Y., Baudin, F., Tafur, L., Murciano, B., Wetzel, R., Weis, F., Muller, C.W.(2019) Nat Commun 10: 5543-5543
- PubMed: 31804486
- DOI: https://doi.org/10.1038/s41467-019-13510-w
- Primary Citation of Related Structures:
6RQH, 6RQL, 6RRD, 6RUI, 6RUO, 6RWE - PubMed Abstract:
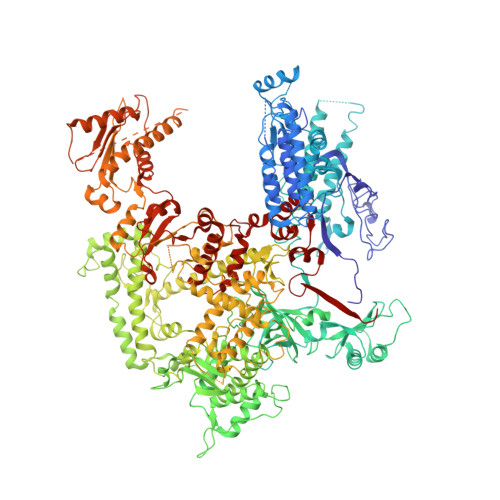
RNA polymerase I (Pol I) assembles with core factor (CF) and Rrn3 on the rDNA core promoter for transcription initiation. Here, we report cryo-EM structures of closed, intermediate and open Pol I initiation complexes from 2.7 to 3.7 Å resolution to visualize Pol I promoter melting and to structurally and biochemically characterize the recognition mechanism of Pol I promoter DNA. In the closed complex, double-stranded DNA runs outside the DNA-binding cleft. Rotation of CF and upstream DNA with respect to Pol I and Rrn3 results in the spontaneous loading and opening of the promoter followed by cleft closure and positioning of the Pol I A49 tandem winged helix domain (tWH) onto DNA. Conformational rearrangement of A49 tWH leads to a clash with Rrn3 to initiate complex disassembly and promoter escape. Comprehensive insight into the Pol I transcription initiation cycle allows comparisons with promoter opening by Pol II and Pol III.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Meyerhofstrasse 1, 69117, Heidelberg, Germany.