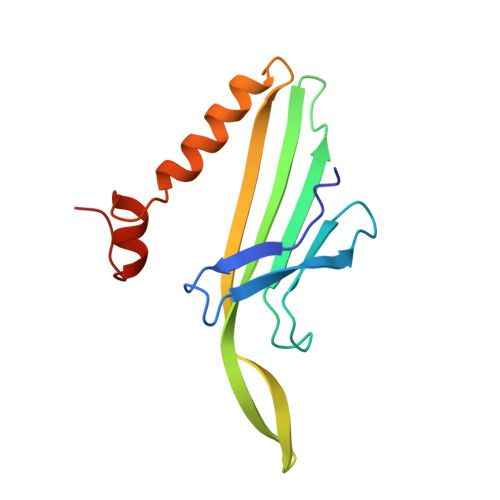
Crystal structures of MS2 capsids with mutations in the subunit FG loop.
Stonehouse, N.J., Valegard, K., Golmohammadi, R., van den Worm, S., Walton, C., Stockley, P.G., Liljas, L.(1996) J Mol Biol 256: 330-339
- PubMed: 8594200
- DOI: https://doi.org/10.1006/jmbi.1996.0089
- Primary Citation of Related Structures:
1BMS, 1MST - PubMed Abstract:
The loop between the F and G beta strands (FG loop) of the bacteriophage MS2 coat protein subunit forms inter-subunit contacts around the 5-fold and 3-fold (quasi 6-fold) axes of the T=3 protein shell. In capsids, the loop is found in two very different conformations, one in B subunits, which form the 5-fold contact, and one in A and C subunits, which form the quasi 6-fold contact. One proline residue, Pro78, is strictly conserved in the coat protein of all related bacteriophages, and in the case of MS2 this proline residue is preceded by a cis peptide bond in the B subunit. In order to probe the role of the FG loop in capsid assembly, we have determined the crystal structures of two MS2 capsids, formed by coat proteins with mutations at two positions in the FG loop, P78N or E76D. These mutants show conformational changes in the FG loops that explain the reduced temperature stability of the capsids. The P78N mutant has a normal trans peptide bond at position 78.
Organizational Affiliation:
Department of Genetics, University of Leeds, UK.