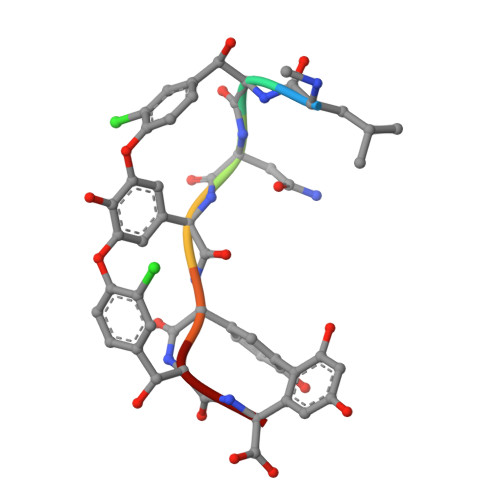
Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Zarkan, A., Macklyne, H.R., Chirgadze, D.Y., Bond, A.D., Hesketh, A.R., Hong, H.J.(2017) Sci Rep 7: 4893-4893
- PubMed: 28687742
- DOI: https://doi.org/10.1038/s41598-017-04868-2
- Primary Citation of Related Structures:
5M2H, 5M2K - PubMed Abstract:
Vancomycin is known to bind to Zn(II) and can induce a zinc starvation response in bacteria. Here we identify a novel polymerization of vancomycin dimers by structural analysis of vancomycin-Zn(II) crystals and fibre X-ray diffraction. Bioassays indicate that this structure is associated with an increased antibiotic activity against bacterial strains possessing high level vancomycin resistance mediated by the reprogramming of peptidoglycan biosynthesis to use precursors terminating in D-Ala-D-Lac in place of D-Ala-D-Ala. Polymerization occurs via interaction of Zn(II) with the N-terminal methylleucine group of vancomycin, and we show that the activity of other glycopeptide antibiotics with this feature can also be similarly augmented by Zn(II). Construction and analysis of a model strain predominantly using D-Ala-D-Lac precursors for peptidoglycan biosynthesis during normal growth supports the hypothesis that Zn(II) mediated vancomycin polymerization enhances the binding affinity towards these precursors.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge, CB2 1QW, UK.