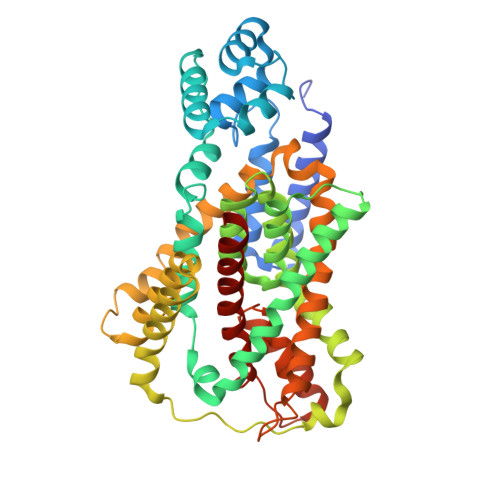
Structural insights into sodium transport by the oxaloacetate decarboxylase sodium pump.
Xu, X., Shi, H., Gong, X., Chen, P., Gao, Y., Zhang, X., Xiang, S.(2020) Elife 9
- PubMed: 32459174
- DOI: https://doi.org/10.7554/eLife.53853
- Primary Citation of Related Structures:
6IVA, 6IWW - PubMed Abstract:
The oxaloacetate decarboxylase sodium pump (OAD) is a unique primary-active transporter that utilizes the free energy derived from oxaloacetate decarboxylation for sodium transport across the cell membrane. It is composed of 3 subunits: the α subunit catalyzes carboxyl-transfer from oxaloacetate to biotin, the membrane integrated β subunit catalyzes the subsequent carboxyl-biotin decarboxylation and the coupled sodium transport, the γ subunit interacts with the α and β subunits and stabilizes the OAD complex. We present here structure of the Salmonella typhimurium OAD βγ sub-complex. The structure revealed that the β and γ subunits form a β 3 γ 3 hetero-hexamer with extensive interactions between the subunits and shed light on the OAD holo-enzyme assembly. Structure-guided functional studies provided insights into the sodium binding sites in the β subunit and the coupling between carboxyl-biotin decarboxylation and sodium transport by the OAD β subunit.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Tianjin Medical University, Tianjin, China.