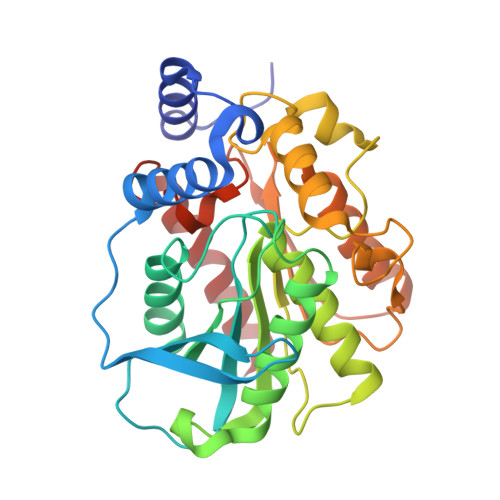
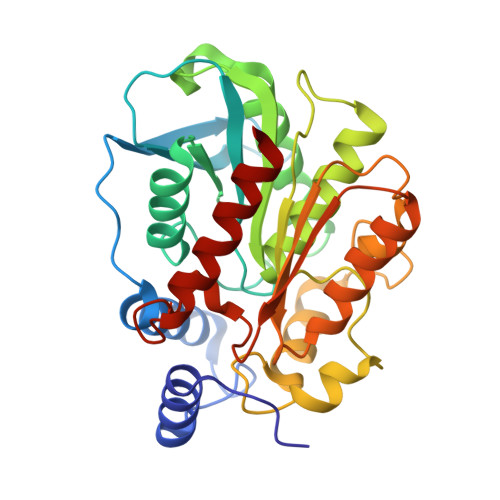
Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
Xiaochen, Y., Yingyi, H., Zhengyang, L., Shuling, J., Zhen, R., Zhao, W., Xiaojian, H., Henglin, C., Jixi, L., Xuewei, X.To be published.