Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
Peng, Q., Peng, R., Qi, J., Gao, G.F., Shi, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polymerase 3 | 710 | Influenza D virus (D/swine/Oklahoma/1334/2011) | Mutation(s): 0 Gene Names: P3 | 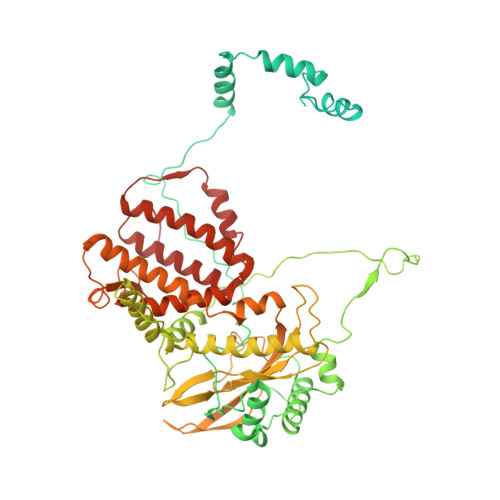 | |
UniProt | |||||
Find proteins for K9LHJ4 (Influenza D virus) Explore K9LHJ4 Go to UniProtKB: K9LHJ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K9LHJ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA-directed RNA polymerase catalytic subunit | 753 | Influenza D virus (D/swine/Oklahoma/1334/2011) | Mutation(s): 0 Gene Names: PB1 EC: 2.7.7.48 | 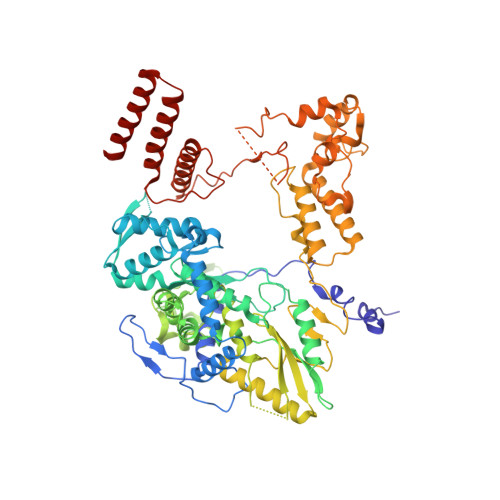 | |
UniProt | |||||
Find proteins for K9LH03 (Influenza D virus) Explore K9LH03 Go to UniProtKB: K9LH03 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K9LH03 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polymerase PB2 | 772 | Influenza D virus (D/swine/Oklahoma/1334/2011) | Mutation(s): 0 Gene Names: PB2 | 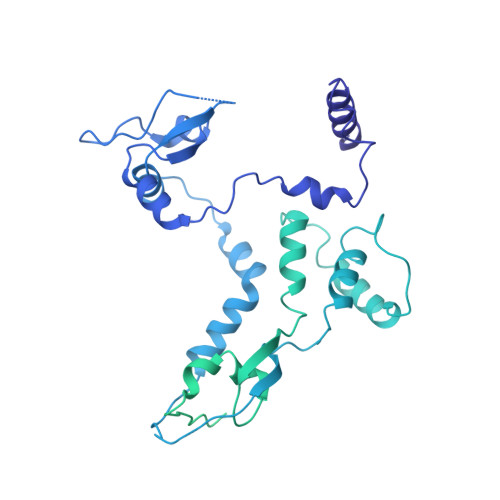 | |
UniProt | |||||
Find proteins for K9LHF3 (Influenza D virus) Explore K9LHF3 Go to UniProtKB: K9LHF3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K9LHF3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 3'-vRNA | D [auth R] | 14 | synthetic construct | 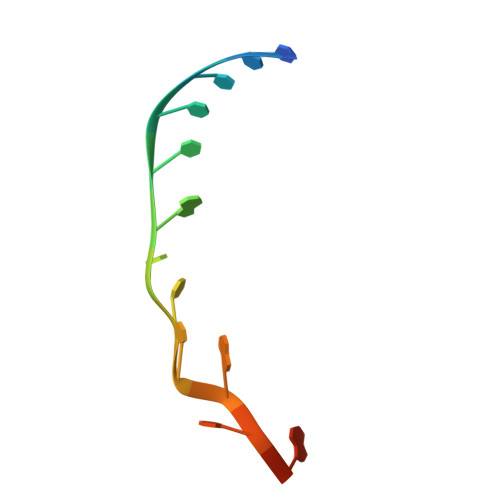 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-vRNA | E [auth V] | 15 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | XDB08020100 |