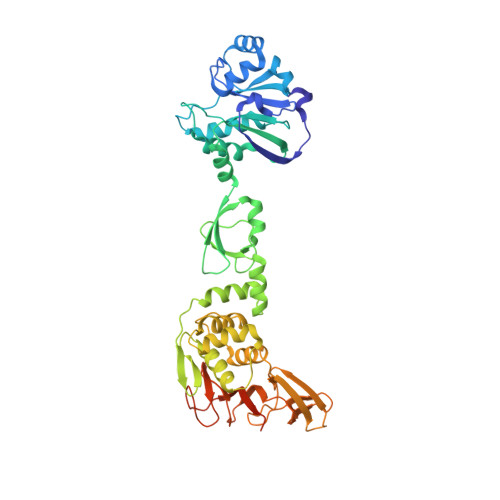
Structure of the multiple functional domains from coronavirus nonstructural protein 3.
Li, M., Ye, G., Si, Y., Shen, Z., Liu, Z., Shi, Y., Xiao, S., Fu, Z.F., Peng, G.(2021) Emerg Microbes Infect 10: 66-80
- PubMed: 33327866
- DOI: https://doi.org/10.1080/22221751.2020.1865840
- Primary Citation of Related Structures:
6LN0 - PubMed Abstract:
Coronaviruses (CoVs) are potential pandemic pathogens that can infect a variety of hosts and cause respiratory, enteric, hepatic and neurological diseases. Nonstructural protein 3 (nsp3), an essential component of the replication/transcription complex, is one of the most important antiviral targets. Here, we report the first crystal structure of multiple functional domains from porcine delta-coronavirus (PDCoV) nsp3, including the macro domain (Macro), ubiquitin-like domain 2 (Ubl2) and papain-like protease (PLpro) catalytic domain. In the asymmetric unit, two of the subunits form the head-to-tail homodimer with an interaction interface between Macro and PLpro. However, PDCoV Macro-Ubl2-PLpro mainly exists as a monomer in solution. Then, we conducted fluorescent resonance energy transfer-based protease assays and found that PDCoV PLpro can cleave a peptide by mimicking the cognate nsp2/nsp3 cleavage site in peptide substrates and exhibits deubiquitinating and de-interferon stimulated gene(deISGylating) activities by hydrolysing ubiquitin-7-amino-4-methylcoumarin (Ub-AMC) and ISG15-AMC substrates. Moreover, the deletion of Macro or Macro-Ubl2 decreased the enzyme activity of PLpro, indicating that Macro and Ubl2 play important roles in maintaining the stability of the PLpro domain. Two active sites of PLpro, Cys260 and His398, were determined; unexpectedly, the conserved site Asp412 was not the third active site. Furthermore, the motif "NGYDT" (amino acids 409-413) was important for stabilizing the enzyme activity of PLpro, and the N409A mutant significantly decreased the enzyme activity of PLpro. These results provide novel insights into the replication mechanism of CoV and new clues for future drug design.
Organizational Affiliation:
State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, People's Republic of China.