6XDG
Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies
- PDB DOI: https://doi.org/10.2210/pdb6XDG/pdb
- EM Map EMD-22137: EMDB EMDataResource
- Classification: VIRAL PROTEIN/IMMUNE SYSTEM
- Organism(s): Severe acute respiratory syndrome coronavirus 2, Homo sapiens
- Expression System: Cricetulus griseus
- Mutation(s): No
- Deposited: 2020-06-10 Released: 2020-06-24
- Funding Organization(s): Biomedical Advanced Research and Development Authority (BARDA)
Experimental Data Snapshot
- Method: ELECTRON MICROSCOPY
- Resolution: 3.90 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
wwPDB Validation 3D Report Full Report
This is version 2.2 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike protein S1 | A [auth E] | 251 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: S, 2 | 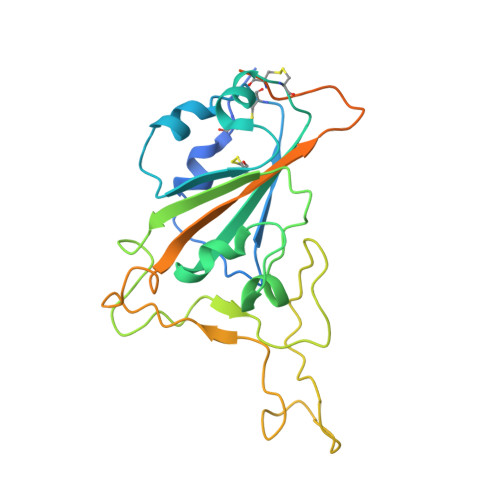 |
UniProt | |||||
Find proteins for P0DTC2 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTC2 Go to UniProtKB: P0DTC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTC2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REGN10933 antibody Fab fragment light chain | B [auth D] | 214 | Homo sapiens | Mutation(s): 0 | 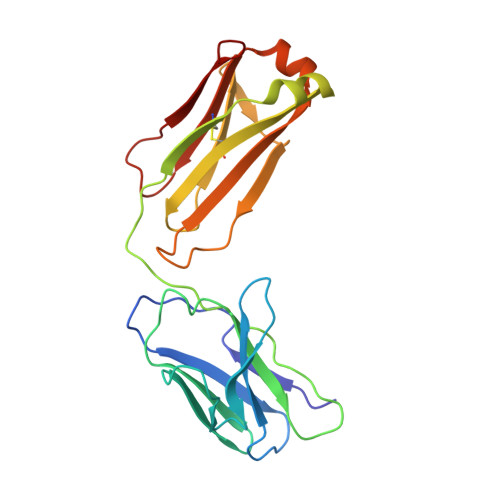 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REGN10933 antibody Fab fragment heavy chain | C [auth B] | 226 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REGN10987 antibody Fab fragment heavy chain | D [auth C] | 225 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REGN10987 antibody Fab fragment light chain | E [auth A] | 216 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | F | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22573RC GlyCosmos: G22573RC GlyGen: G22573RC | |||||
Experimental Data & Validation
Experimental Data
- Method: ELECTRON MICROSCOPY
- Resolution: 3.90 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | |
| MODEL REFINEMENT | PHENIX |
Entry History & Funding Information
Deposition Data
- Released Date: 2020-06-24 Deposition Author(s): Franklin, M.C., Saotome, K., Romero Hernandez, A., Zhou, Y.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biomedical Advanced Research and Development Authority (BARDA) | HHSO100201700020C |
Revision History (Full details and data files)
- Version 1.0: 2020-06-24
Type: Initial release - Version 1.1: 2020-07-08
Changes: Author supporting evidence - Version 1.2: 2020-07-22
Changes: Source and taxonomy - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Advisory, Atomic model, Data collection, Derived calculations, Structure summary - Version 2.1: 2020-09-02
Changes: Database references, Structure summary - Version 2.2: 2021-01-27
Changes: Structure summary













