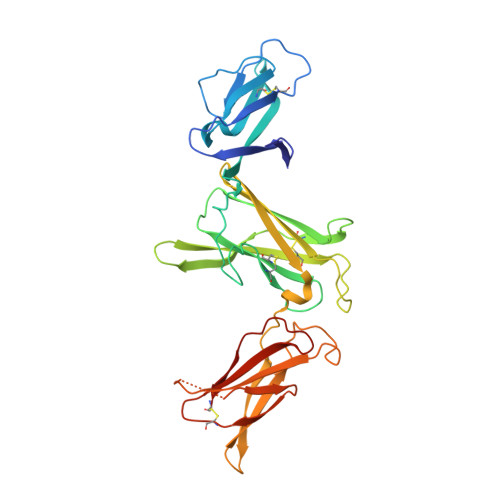
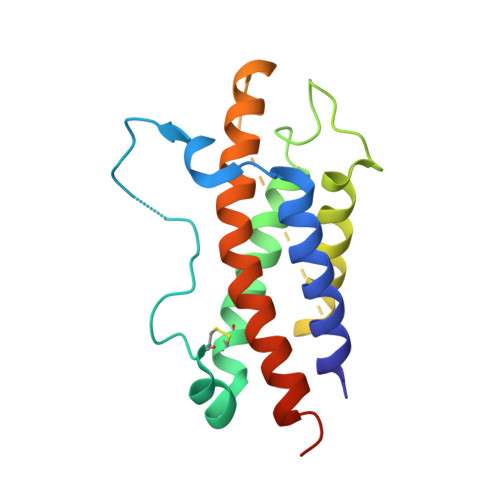
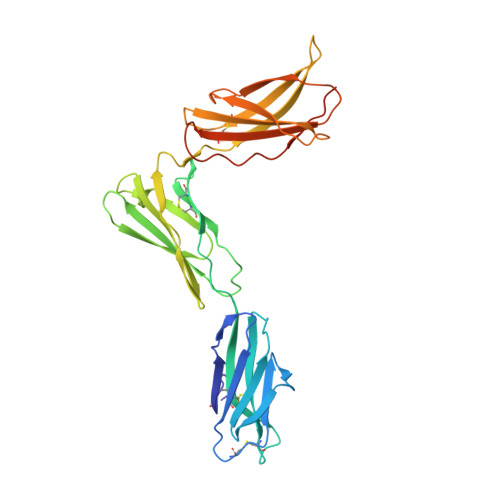
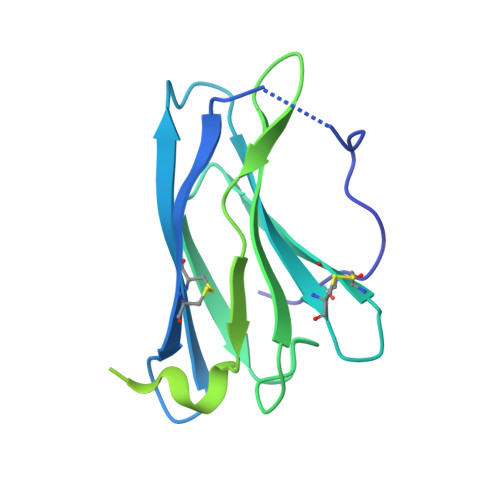
Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Glassman, C.R., Mathiharan, Y.K., Jude, K.M., Su, L., Panova, O., Lupardus, P.J., Spangler, J.B., Ely, L.K., Thomas, C., Skiniotis, G., Garcia, K.C.(2021) Cell 184: 983-999.e24
- PubMed: 33606986
- DOI: https://doi.org/10.1016/j.cell.2021.01.018
- Primary Citation of Related Structures:
6WDP, 6WDQ - PubMed Abstract:
Interleukin-12 (IL-12) and IL-23 are heterodimeric cytokines that are produced by antigen-presenting cells to regulate the activation and differentiation of lymphocytes, and they share IL-12Rβ1 as a receptor signaling subunit. We present a crystal structure of the quaternary IL-23 (IL-23p19/p40)/IL-23R/IL-12Rβ1 complex, together with cryoelectron microscopy (cryo-EM) maps of the complete IL-12 (IL-12p35/p40)/IL-12Rβ2/IL-12Rβ1 and IL-23 receptor (IL-23R) complexes, which reveal "non-canonical" topologies where IL-12Rβ1 directly engages the common p40 subunit. We targeted the shared IL-12Rβ1/p40 interface to design a panel of IL-12 partial agonists that preserved interferon gamma (IFNγ) induction by CD8 + T cells but impaired cytokine production from natural killer (NK) cells in vitro. These cell-biased properties were recapitulated in vivo, where IL-12 partial agonists elicited anti-tumor immunity to MC-38 murine adenocarcinoma absent the NK-cell-mediated toxicity seen with wild-type IL-12. Thus, the structural mechanism of receptor sharing used by IL-12 family cytokines provides a protein interface blueprint for tuning this cytokine axis for therapeutics.
Organizational Affiliation:
Program in Immunology, Stanford University School of Medicine, Stanford, CA 94305, USA; Departments of Molecular and Cellular Physiology and Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.