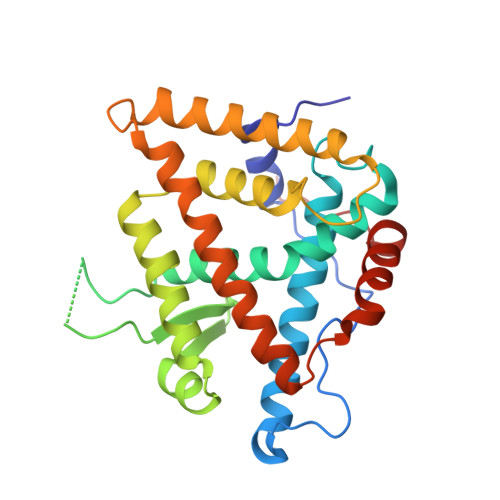
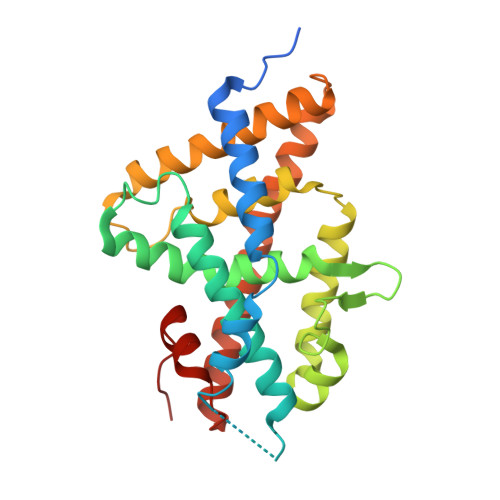
Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
Browning, C., McEwen, A.G., Mori, K., Yokoi, T., Moras, D., Nakagawa, Y., Billas, I.M.L.(2021) J Pestic Sci 46: 88-100
- PubMed: 33746550
- DOI: https://doi.org/10.1584/jpestics.D20-095
- Primary Citation of Related Structures:
7BJU, 7BJV - PubMed Abstract:
The ecdysone receptor (EcR) possesses the remarkable capacity to adapt structurally to different types of ligands. EcR binds ecdysteroids, including 20-hydroxyecdysone (20E), as well as nonsteroidal synthetic agonists such as insecticidal dibenzoylhydrazines (DBHs). Here, we report the crystal structures of the ligand-binding domains of Heliothis virescens EcR/USP bound to the DBH agonist BYI09181 and to the imidazole-type compound BYI08346. The region delineated by helices H7 and H10 opens up to tightly fit a phenyl ring of the ligands to an extent that depends on the bulkiness of ring substituent. In the structure of 20E-bound EcR, this part of the ligand-binding pocket (LBP) contains a channel filled by water molecules that form an intricate hydrogen bond network between 20E and LBP. The water channel present in the nuclear receptor bound to its natural hormone acts as a critical molecular adaptation spring used to accommodate synthetic agonists inside its binding cavity.
Organizational Affiliation:
Centre for Integrative Biology (CBI), Department of Integrated Structural Biology, IGBMC (Institute of Genetics and of Molecular and Cellular Biology), Illkirch, France.