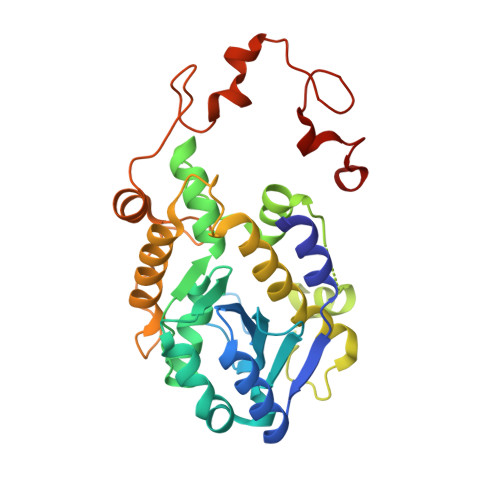
Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Gao, Y., Liu, H., Zhang, C., Su, S., Chen, Y., Chen, X., Li, Y., Shao, Z., Zhang, Y., Shao, Q., Li, J., Huang, Z., Ma, J., Gan, J.(2021) Nucleic Acids Res 49: 568-583
- PubMed: 33332555
- DOI: https://doi.org/10.1093/nar/gkaa1197
- Primary Citation of Related Structures:
7C42, 7C43, 7C45, 7C47, 7C4B, 7C4C - PubMed Abstract:
Infection with kinetoplastid parasites, including Trypanosoma brucei (T. brucei), Trypanosoma cruzi (T. cruzi) and Leishmania can cause serious disease in humans. Like other kinetoplastid species, mRNAs of these disease-causing parasites must undergo posttranscriptional editing in order to be functional. mRNA editing is directed by gRNAs, a large group of small RNAs. Similar to mRNAs, gRNAs are also precisely regulated. In T. brucei, overexpression of RNase D ribonuclease (TbRND) leads to substantial reduction in the total gRNA population and subsequent inhibition of mRNA editing. However, the mechanisms regulating gRNA binding and cleavage by TbRND are not well defined. Here, we report a thorough structural study of TbRND. Besides Apo- and NMP-bound structures, we also solved one TbRND structure in complexed with single-stranded RNA. In combination with mutagenesis and in vitro cleavage assays, our structures indicated that TbRND follows the conserved two-cation-assisted mechanism in catalysis. TbRND is a unique RND member, as it contains a ZFD domain at its C-terminus. In addition to T. brucei, our studies also advanced our understanding on the potential gRNA degradation pathway in T. cruzi, Leishmania, as well for as other disease-associated parasites expressing ZFD-containing RNDs.
Organizational Affiliation:
Shanghai Public Health Clinical Center, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Department of Physiology and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.