Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Wang, K., Zheng, B., Zhang, L., Cui, L., Su, X., Zhang, Q., Guo, Z., Guo, Y., Zhang, W., Zhu, L., Zhu, F., Rao, Z., Wang, X.(2020) Nat Commun 11: 4419-4419
- PubMed: 32887892
- DOI: https://doi.org/10.1038/s41467-020-18250-w
- Primary Citation of Related Structures:
7C80, 7C81, 7CMK - PubMed Abstract:
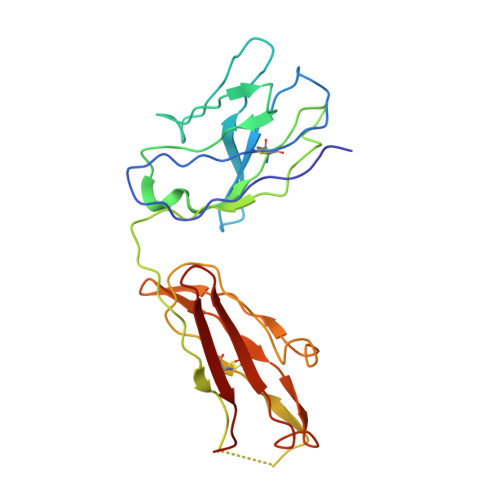
Echovirus 30 (E30), a serotype of Enterovirus B (EV-B), recently emerged as a major causative agent of aseptic meningitis worldwide. E30 is particularly devastating in the neonatal population and currently no vaccine or antiviral therapy is available. Here we characterize two highly potent E30-specific monoclonal antibodies, 6C5 and 4B10, which efficiently block binding of the virus to its attachment receptor CD55 and uncoating receptor FcRn. Combinations of 6C5 and 4B10 augment the sum of their individual anti-viral activities. High-resolution structures of E30-6C5-Fab and E30-4B10-Fab define the location and nature of epitopes targeted by the antibodies. 6C5 and 4B10 engage the capsid loci at the north rim of the canyon and in-canyon, respectively. Notably, these regions exhibit antigenic variability across EV-Bs, highlighting challenges in development of broad-spectrum antibodies. Our structures of these neutralizing antibodies of E30 are instructive for development of vaccines and therapeutics against EV-B infections.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences and College of Pharmacy and Drug Discovery Center for Infectious Diseases, Nankai University, 300353, Tianjin, China.