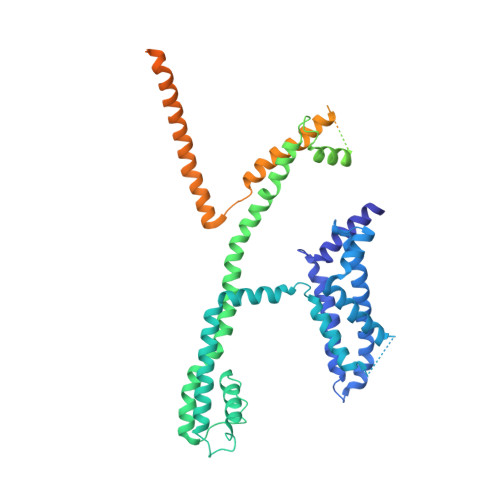
Molecular basis for ligand activation of the human KCNQ2 channel.
Li, X., Zhang, Q., Guo, P., Fu, J., Mei, L., Lv, D., Wang, J., Lai, D., Ye, S., Yang, H., Guo, J.(2021) Cell Res 31: 52-61
- PubMed: 32884139
- DOI: https://doi.org/10.1038/s41422-020-00410-8
- Primary Citation of Related Structures:
7CR0, 7CR1, 7CR2, 7CR3, 7CR4, 7CR7 - PubMed Abstract:
The voltage-gated potassium channel KCNQ2 is responsible for M-current in neurons and is an important drug target to treat epilepsy, pain and several other diseases related to neuronal hyper-excitability. A list of synthetic compounds have been developed to directly activate KCNQ2, yet our knowledge of their activation mechanism is limited, due to lack of high-resolution structures. Here, we report cryo-electron microscopy (cryo-EM) structures of the human KCNQ2 determined in apo state and in complex with two activators, ztz240 or retigabine, which activate KCNQ2 through different mechanisms. The activator-bound structures, along with electrophysiology analysis, reveal that ztz240 binds at the voltage-sensing domain and directly stabilizes it at the activated state, whereas retigabine binds at the pore domain and activates the channel by an allosteric modulation. By accurately defining ligand-binding sites, these KCNQ2 structures not only reveal different ligand recognition and activation mechanisms, but also provide a structural basis for drug optimization and design.
Organizational Affiliation:
Department of Biophysics, and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310058, China.