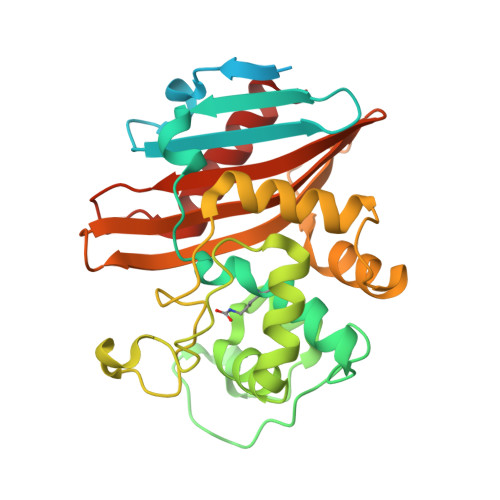
Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Stewart, N.K., Toth, M., Stasyuk, A., Lee, M., Smith, C.A., Vakulenko, S.B.(2021) ACS Infect Dis 7: 1164-1176
- PubMed: 33390002
- DOI: https://doi.org/10.1021/acsinfecdis.0c00714
- Primary Citation of Related Structures:
7KEP, 7KEQ, 7KER - PubMed Abstract:
Avibactam is a potent diazobicyclooctane inhibitor of class A and C β-lactamases. The inhibitor also exhibits variable activity against some class D enzymes from Gram-negative bacteria; however, its interaction with recently discovered class D β-lactamases from Gram-positive bacteria has not been studied. Here, we describe microbiological, kinetic, and mass spectrometry studies of the interaction of avibactam with CDD-1, a class D β-lactamase from the clinically important pathogen Clostridioides difficile , and show that avibactam is a potent irreversible mechanism-based inhibitor of the enzyme. X-ray crystallographic studies at three time-points demonstrate the rapid formation of a stable CDD-1-avibactam acyl-enzyme complex and highlight differences in the anchoring of the inhibitor by class D enzymes from Gram-positive and Gram-negative bacteria.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, United States.