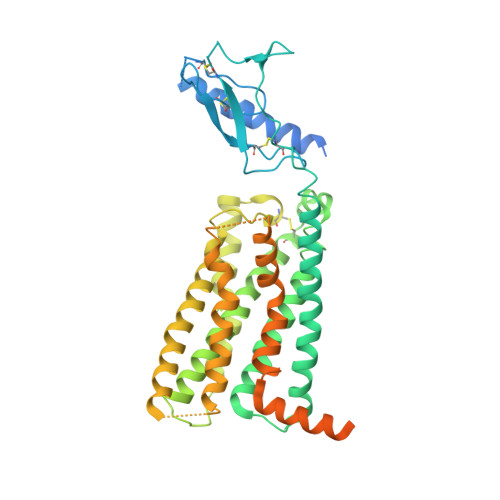
Structure and dynamics of the CGRP receptor in apo and peptide-bound forms.
Josephs, T.M., Belousoff, M.J., Liang, Y.L., Piper, S.J., Cao, J., Garama, D.J., Leach, K., Gregory, K.J., Christopoulos, A., Hay, D.L., Danev, R., Wootten, D., Sexton, P.M.(2021) Science 372
- PubMed: 33602864
- DOI: https://doi.org/10.1126/science.abf7258
- Primary Citation of Related Structures:
7KNT, 7KNU - PubMed Abstract:
G protein-coupled receptors (GPCRs) are key regulators of information transmission between cells and organs. Despite this, we have only a limited understanding of the behavior of GPCRs in the apo state and the conformational changes upon agonist binding that lead to G protein recruitment and activation. We expressed and purified unmodified apo and peptide-bound calcitonin gene-related peptide (CGRP) receptors from insect cells to determine their cryo-electron microscopy (cryo-EM) structures, and we complemented these with analysis of protein conformational dynamics using hydrogen-deuterium exchange mass spectrometry and three-dimensional variance analysis of the cryo-EM data. Together with our previously published structure of the active, Gs-bound CGRP receptor complex, our work provides insight into the mechanisms of class B1 GPCR activation.
Organizational Affiliation:
Drug Discovery Biology Theme, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, Victoria, Australia.