Full Text |
QUERY: Scientific Name of the Source Organism = "Haloferax volcanii" | MyPDB Login | Search API |
| Search Summary | This query matches 12 Polymer Entities. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 12 of 12 Polymer Entities Page 1 of 1 Sort by
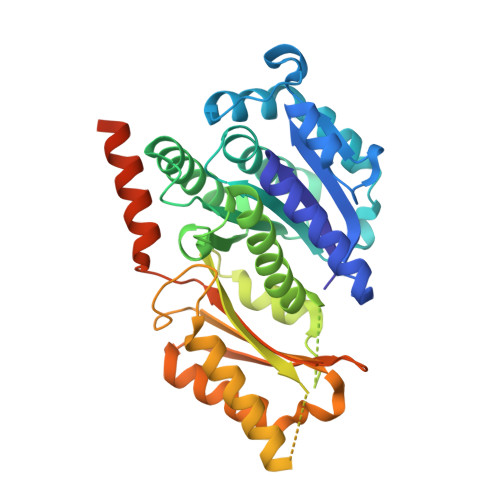
DIHYDROFOLATE REDUCTASE(1998) Structure 6: 75-88
NMR Structure of Haloferax volcanii DHFR(2007) Protein Sci 16: 1783-1787
binary hvDHFR1:folate complexBoroujerdi, A.A.F.B., Young, J.K. (2009) Biopolymers 91: 140-144
Crystal structure of proliferating cell nuclear antigen (PCNA) from Haloferax volcaniiMorgunova, E., Gray, F.C., MacNeill, S.A., Ladenstein, R. (2009) Acta Crystallogr D Biol Crystallogr 65: 1081-1088
Crystal structure of the Haloferax volcanii proliferating cell nuclear antigenWinter, J.A., Christofi, P., Morroll, S., Bunting, K.A. (2009) BMC Struct Biol 9: 55-55
CetZ2 from Haloferax volcanii - GTPgS bound protofilamentAylett, C.H.S., Duggin, I.G., Lowe, J. (2015) Nature 519: 362
CetZ2 from Haloferax volcanii - GTPgS bound protofilamentAylett, C.H.S., Duggin, I.G., Lowe, J. (2015) Nature 519: 362
CetZ1 from Haloferax volcanii - GDP bound monomerAylett, C.H.S., Duggin, I.G., Lowe, J. (2015) Nature 519: 362
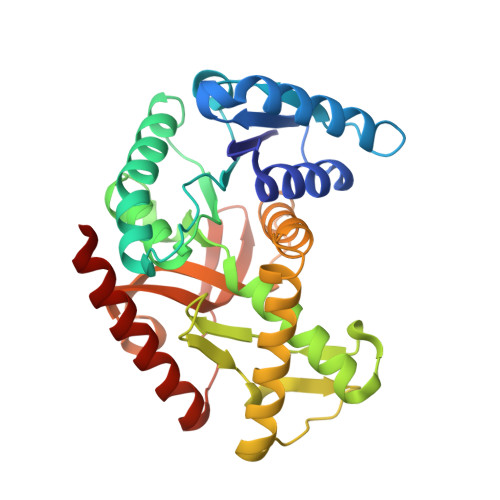
1.50 A resolution structure of the malate dehydrogenase from Haloferax volcaniiTalon, R., Madern, D., Girard, E. To be published
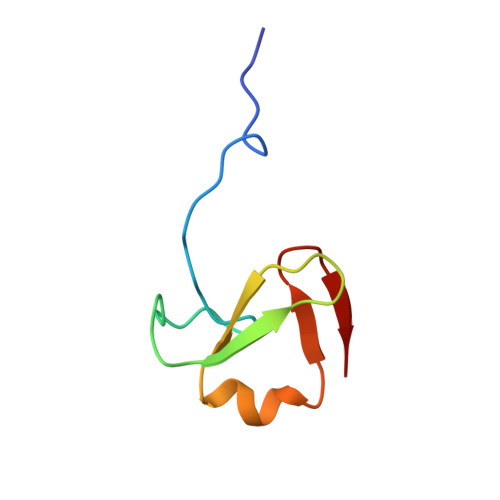
Solution structure and dynamics of Zn-Finger HVO_2753 proteinKubatova, N., Pyper, D., Schwalbe, H. To be published
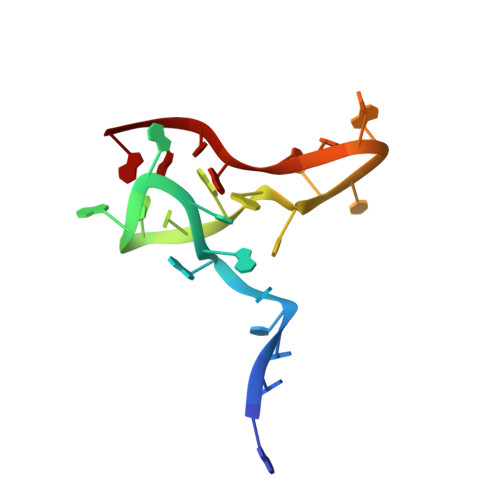
Structural basis of translation inhibition by a valine tRNA-derived fragment(2024) Life Sci Alliance 7:
Structural basis of translation inhibition by a valine tRNA-derived fragment(2024) Life Sci Alliance 7:
1 to 12 of 12 Polymer Entities Page 1 of 1 Sort by |