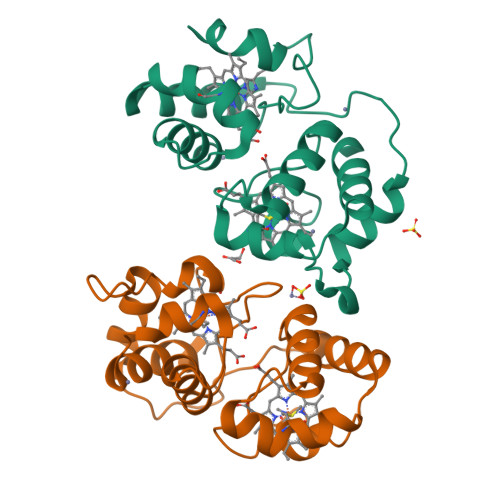
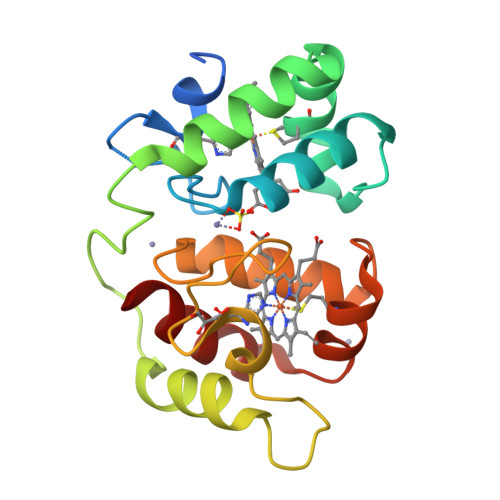
The Structure of Acidithiobacillus Ferrooxidans C(4)-Cytochrome. A Model for Complex-Induced Electron Transfer Tuning
Abergel, C., Nitschke, W., Malarte, G., Bruschi, M., Claverie, J.-M., Guidici-Orticoni, M.-T.(2003) Structure 11: 547
- PubMed: 12737820
- DOI: https://doi.org/10.1016/s0969-2126(03)00072-8
- Primary Citation of Related Structures:
1H1O - PubMed Abstract:
The study of electron transfer between the copper protein rusticyanin (RCy) and the c(4)-cytochrome CYC(41) of the acidophilic bacterium Acidithiobacillus ferrooxidans has evidenced a remarkable decrease of RCy's redox potential upon complex formation. The structure of the CYC(41) obtained at 2.2 A resolution highlighted a specific glutamate residue (E121) involved in zinc binding as potentially playing a central role in this effect, required for the electron transfer to occur. EPR and stopped-flow experiments confirmed the strong inhibitory effect of divalent cations on CYC(41):RCy complex formation. A docking analysis of the CYC(41) and RCy structure allows us to propose a detailed model for the complex-induced tuning of electron transfer in agreement with our experimental data, which could be representative of other copper proteins involved in electron transfer.
Organizational Affiliation:
Information Génomique et Structurale, UPR 2589, Institut de Biologie Structurale et Microbiologie, 31 Chemin Joseph Aiguier, 13402 20, Marseille cedex, France. chantal.abergel@igs.cnrs-mrs.fr